Chapter 15 Regularization and Feature Selection
We assume you have loaded the following packages:
Below we load more as we introduce more.
For replicability, we also set the seed:
15.2 Forward selection
There are several solutions to this problem. A popular algorithm is forward selection where one first picks the best 1-feature model, thereafter tries adding all remaining features one-by-one to build the best two-feature model, and thereafter the best three-feature model, and so on, until the model performance starts to deteriorate.
0-feature base model:
m0 = LinearRegression(fit_intercept = False)
X0 = np.ones((X.shape[0], 1))
X0t, X0v, yt, yv = train_test_split(X0, y, test_size = 0.4,
random_state = 8)
_t = m0.fit(X0t, yt)
m0.score(X0v, yv)## -0.006849647990198937Try 1-feature models:
best1 = {"i": -1, "R2": -np.inf}
m1 = LinearRegression(fit_intercept = True)
for i in range(X.shape[1]):
X1 = X.iloc[:,[i]]
X1t, X1v, yt, yv = train_test_split(X1, y, test_size = 0.4,
random_state = 8)
_t = m1.fit(X1t, yt)
s = m1.score(X1v, yv)
if s > best1["R2"]:
best1["i"] = i
best1["R2"] = s
best1## {'i': 9, 'R2': 0.4666428183761875}Add another feature:
best2 = {"i": -1, "R2": -np.inf}
m2 = LinearRegression(fit_intercept = True)
included_features = set([best1["i"]])
features_to_test = set(range(X.shape[1])) - included_features
for f in features_to_test:
features = included_features.union({f})
X2 = X.iloc[:, list(features)]
X2t, X2v, yt, yv = train_test_split(X2, y, test_size = 0.4,
random_state = 8)
_t = m2.fit(X2t, yt)
s = m2.score(X2v, yv)
if s > best2["R2"]:
best2["i"] = f
best2["R2"] = s
best2## {'i': 2, 'R2': 0.8677935861014889}Can add more in this fashion.
Make a function:
def bestFeature(X, y, included_features):
best = {"i": -1, "R2": -np.inf}
features_to_test = list(
set(range(X.shape[1])) - set(included_features))
for f in features_to_test:
features = included_features + [f]
Xf = X.iloc[:, features]
Xft, Xfv, yt, yv = train_test_split(Xf, y, test_size = 0.4,
random_state = 8)
_t = m2.fit(Xft, yt)
s = m2.score(Xfv, yv)
if s > best["R2"]:
best["i"] = f
best["R2"] = s
return best
best = list()
included_features = []
r2s = []
for f in range(X.shape[1]):
b = bestFeature(X, y, included_features)
best.append(b)
included_features = included_features + [b["i"]]
r2s.append(b["R2"])
best## [{'i': 9, 'R2': 0.4666428183761875}, {'i': 2, 'R2': 0.8677935861014889}, {'i': 27, 'R2': 0.9296522813032738}, {'i': 28, 'R2': 0.9396723227644169}, {'i': 14, 'R2': 0.9430611422142994}, {'i': 22, 'R2': 0.9437993501430608}, {'i': 33, 'R2': 0.9446022123630592}, {'i': 59, 'R2': 0.9450369454683462}, {'i': 10, 'R2': 0.9452498568205782}, {'i': 8, 'R2': 0.9455986815032276}, {'i': 19, 'R2': 0.9462862467428159}, {'i': 1, 'R2': 0.9464320773146165}, {'i': 46, 'R2': 0.9465914853888853}, {'i': 40, 'R2': 0.9465889082229213}, {'i': 55, 'R2': 0.9465359196145449}, {'i': 36, 'R2': 0.9464542982901596}, {'i': 5, 'R2': 0.9463762822853345}, {'i': 20, 'R2': 0.9462833979835922}, {'i': 34, 'R2': 0.9462050456158057}, {'i': 48, 'R2': 0.9461478229257206}, {'i': 50, 'R2': 0.9461054197700751}, {'i': 52, 'R2': 0.9460730478692239}, {'i': 26, 'R2': 0.9459904691472554}, {'i': 42, 'R2': 0.9459265694540916}, {'i': 44, 'R2': 0.9458087497124463}, {'i': 32, 'R2': 0.9455590142220006}, {'i': 39, 'R2': 0.945408502141411}, {'i': 25, 'R2': 0.9449675269753557}, {'i': 24, 'R2': 0.9445346914386826}, {'i': 58, 'R2': 0.9438665545550707}, {'i': 54, 'R2': 0.9431161004717253}, {'i': 38, 'R2': 0.9425908604390918}, {'i': 21, 'R2': 0.9418808150038418}, {'i': 35, 'R2': 0.9414073375383587}, {'i': 37, 'R2': 0.9410776069690762}, {'i': 41, 'R2': 0.940836636399586}, {'i': 49, 'R2': 0.9406534112839524}, {'i': 51, 'R2': 0.940509615245991}, {'i': 53, 'R2': 0.9403938540834724}, {'i': 43, 'R2': 0.9401119255355864}, {'i': 57, 'R2': 0.9397885757150655}, {'i': 23, 'R2': 0.9394085387576068}, {'i': 12, 'R2': 0.9379313435724337}, {'i': 30, 'R2': 0.9377835490332745}, {'i': 45, 'R2': 0.9333091167915868}, {'i': 29, 'R2': 0.9340676090843362}, {'i': 16, 'R2': 0.9285874234675286}, {'i': 0, 'R2': 0.9182942310441039}, {'i': 6, 'R2': 0.880809369532485}, {'i': 56, 'R2': 0.8437760019671372}, {'i': 31, 'R2': 0.8474899063079687}, {'i': 47, 'R2': 0.8521445384395225}, {'i': 17, 'R2': -9.818297650553768e+22}, {'i': 13, 'R2': -2.000001247173613e+22}, {'i': 15, 'R2': -4.135131612681403e+22}, {'i': 3, 'R2': -2.7600188239712023e+22}, {'i': 4, 'R2': -1.1141317933826631e+23}, {'i': 7, 'R2': -5.35812572220869e+22}, {'i': 18, 'R2': -3.2569968014607136e+22}, {'i': 11, 'R2': -3.4476870254964047e+22}]## (-0.1, 1.0)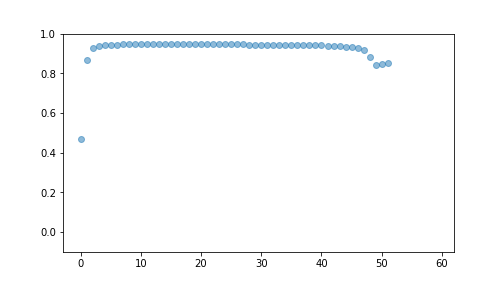
plot of chunk unnamed-chunk-10
from sklearn.feature_selection import SequentialFeatureSelector
m = LinearRegression()
sfs = SequentialFeatureSelector(m)
_t = sfs.fit(Xt, yt)
sfs.get_feature_names_out()## array(['x3', 'x10', 'z1_B', 'z1_C', 'z2_B', 'z2_C', 'z4_B', 'z4_C',
## 'z5_B', 'z6_B', 'z7_B', 'z7_C', 'z8_B', 'z10_B', 'z10_C', 'z12_B',
## 'z12_C', 'z13_B', 'z13_C', 'z14_B', 'z14_C', 'z15_B', 'z15_C',
## 'z16_B', 'z17_B', 'z18_B', 'z18_C', 'z19_B', 'z20_B', 'z20_C'],
## dtype=object)## 0.932918373971302515.3 Ridge and Lasso regression
Ridge and Lasso are methods that are related to forward selection. These methods penalize large \(\beta\) values and hence suppress or eliminate correlated variables. These do not need looping over different combinations of variables like forward selection, however, one normally has to loop over the penalty parameter alpha to find the optimal value.
Both methods are living in sklearn.linear_regression.
We demonstrate this with ridge regression below:
from sklearn.linear_model import Ridge, Lasso
m = Ridge(alpha = 1) # penalty value 1
cross_val_score(m, X, y, cv=10).mean()## NameError: name 'cross_val_score' is not defined## NameError: name 'cross_val_score' is not definedAs evident from the above, penalty values 1 and 0.3 give virtually equal results. The cross-validated \(R^2\) is also comparable to what forward-selection suggested.
Lasso works in an analogous fashion as ridge, but as it’s penalty is not globally differentiable (it is \(L_1\) penalty), then you may run into more problems with convergence.
Both methods have more options, in particular they can normalize the features before fitting.
## 0.9264571331356263## 0.8627943726880424