Chapter 10 Linear Regression
We assume you have loaded the following packages:
Below we load more as we introduce more.
10.1 Solving Linear Regression Tasks Manually
10.1.1 The task
In case of simple regression, the task is to find parameters \(\beta_0\) and \(\beta_1\) such that the mean squared error (MSE) is minimized wher MSE is defined as \[\begin{equation} MSE = \frac{1}{n}\sum_i (y_i - \hat y(x_i))^2 \end{equation}\] where the predicted value \[\begin{equation} \hat y(x_i) = \beta_0 + \beta_1\cdot x_i. \end{equation}\] Here \(n\) is the number of observations, \(x\) is our exogenous variable, and \(y\) is the outcome variable, and \(i\) indexes the observations. Normally we want to use software to perform this optimization (even more, this problem can be solved analytically) but it is instructive to attempt to solve the problem by hand.
Let us experiment with iris data and estimate the relationship between petal width and length of versicolor flowers. This is one of the most popular statistics and machine learning dataset, the version we use here originates from R datasets. You can download it from the Bitbucket repo of these notes. The dataset itself contains three species, and as their leaves may have different relationship, we filter out only one of those, versicolor. Also, as the variable names in this file are not suitable for modeling below, we rename variable called Petal.Length to plength, and Petal.Width to pwidth.
First we load the data, and thereafter filter with rename chained at the end of the filtering operation:
iris = pd.read_csv("../data/iris.csv.bz2", sep="\t")
# select only versicolor species,
# make variable names easier to handle
versicolor = iris[iris.Species == "versicolor"].rename(
columns={"Petal.Length": "plength", "Petal.Width":"pwidth"})
# check if data looks reasonable
versicolor.head(3)## Sepal.Length Sepal.Width plength pwidth Species
## 50 7.0 3.2 4.7 1.4 versicolor
## 51 6.4 3.2 4.5 1.5 versicolor
## 52 6.9 3.1 4.9 1.5 versicolorplt.scatter(versicolor.plength, versicolor.pwidth, edgecolor="k")
plt.xlabel("Petal length (cm)")
plt.ylabel("Petal width (cm)")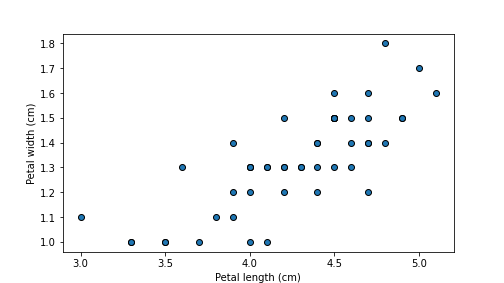
plot of chunk plotIris
The plot reveals that lengths and widths are positively correlated. This is to be expected, longer leaves are wider too.
10.1.2 Solve it manually
Denote petal length by \(l\) and petal width by \(w\). Solving the problem manually involves the following steps:
- Pick suitable values for \(\beta_0\) and \(\beta_1\).
- Compute the predicted petal width, \(\hat w_i = \beta_0 + \beta_1\cdot l_i\).
- Computing the deviations \(e_i = w_i - \hat w_i\).
- Compute MSE as \(MSE = \frac{1}{N} \sum_i e_i^2\).
Let us first write standalone code to perform these steps, and thereafter put all this into a function.
- For a start, we can pick \(\beta_0 = 0\) and \(\beta_1 = 1\). This would amount to quadratic (or circular) leaves:
- Now we compute the predicted values. Remember, the petal length is
called
plengthand petal widht is calledpwin theversicolordataframe:
hat_w = beta0 + beta1*versicolor.plength # note: vectorized operators!
hat_w.head(3) # this is a series!## 50 4.7
## 51 4.5
## 52 4.9
## Name: plength, dtype: float64The results are too wide–the true values are roughly 1.5, not 4.5. Apparently the choice of \(\beta_0\) and \(\beta_1\) was not a good one.
- Compute deviations \(e_i\):
## 50 -3.3
## 51 -3.0
## 52 -3.4
## dtype: float64- Finally, compute MSE. Note that we can use the built-in
np.meanto achieve this:
## 8.7198We get an \(MSE \approx 8.72\). This number in isolation does not tell much, but one can pick a different set of parameters \(\beta_0\) and \(\beta_1\), and compare the resulting MSE-s. This allows us to tell which choice is better.
10.1.3 Create a function to make it more compact
Now we can wrap these steps into a function to make the code more compact. The function might take two arguments, \(\beta_0\) and \(\beta_1\), so we can easily play with different values:
def mse(beta0, beta1):
hat_w = beta0 + beta1*versicolor.plength # note: vectorized operators!
e = versicolor.pwidth - hat_w
mse = np.mean(e**2)
return mseNow we can quickly try a few different parameter values to find out which combination works better:
## 8.7198## 0.6672## 0.11320000000000001So out of these three attempts, the best parameter values are \(\beta_0 = -0.5\) and \(\beta_1 = 0.5\).
10.1.4 Visualize the Regression Line
So far we just computed \(MSE\) but we can do even better–we can also visualize the regression line. Matplotlib does not have a ready-made function to plot a regression line but it is easy to do by hand. We just create a number of sepal length values in a suitable range, predict the corresponding widths, and plot the results.
beta0, beta1 = -0.5, 0.5
# start by plotting the data
plt.scatter(versicolor.plength, versicolor.pwidth, edgecolor="k")
# create 11 evenly spaced lengths between the smallest and largest
# value in the data
length = np.linspace(versicolor.plength.min(), versicolor.plength.max(), 11)
# predict width
hatw = beta0 + beta1*length
plt.plot(length, hatw, c='r')
plt.xlabel("Petal length")
plt.ylabel("Petal width")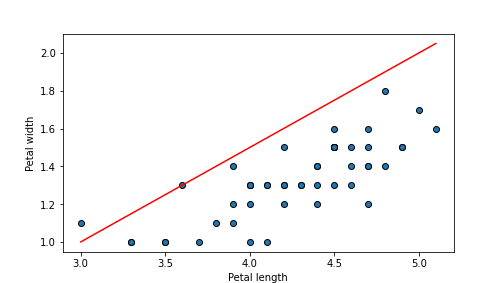
plot of chunk regressionLine
Despite the impressive \(MSE\) value, the figure shows that our line does not actually align well with data. We might continue trying different values to find an even better one.
Exercise 10.1 Write a function that takes \(\beta_0\) and \(\beta_1\) as arguments and a) computes \(MSE\) and b) plot the regression line. Experiment with the parameter values and find a combination that makes the line align well with the data points.
See the solution
10.2 Linear Regression in python: statsmodels.formula.api and sklearn
We discuss two popular libraries for doing linear regression in
python. The first one, statsmodels.formula.api is useful if we want
to interpret the model coefficients, explore \(t\)-values, and assess
the overall model goodness. It is based on R-style
formulas, and it provides well-designed summary tables. The other,
sklearn, is more suitable for predictive modeling. Instead of
formula it expects the design matrix, and it does not provide nicely
formatted summary tables. However, it can be easily swapped with
other predictive models in sklearn module.
10.2.1 Statsmodels and its formula API
We import the statsmodels formula API functionality as
It provides a number of statistical models, include ols, ordinary
least squares, i.e. linear regression. The basic syntax of ols is
The formula is an R-style formula, such as "y ~ x1 + x2" and data is a data frame that contains the variables. For
instance, the versicolor petal regression can be done as
This line just sets up the linear regression model but does not
actually evaluate it, in order to get an answer, one has to add .fit():
This results in evaluated model object m that provides several
useful methods, in particular .params for parameter estimates, and
.summary() for a summary table:
## Intercept -0.084288
## plength 0.331054
## dtype: float64| Dep. Variable: | pwidth | R-squared: | 0.619 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.611 |
| Method: | Least Squares | F-statistic: | 77.93 |
| Date: | Tue, 13 Jan 2026 | Prob (F-statistic): | 1.27e-11 |
| Time: | 17:00:33 | Log-Likelihood: | 34.709 |
| No. Observations: | 50 | AIC: | -65.42 |
| Df Residuals: | 48 | BIC: | -61.59 |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -0.0843 | 0.161 | -0.525 | 0.602 | -0.407 | 0.239 |
| plength | 0.3311 | 0.038 | 8.828 | 0.000 | 0.256 | 0.406 |
| Omnibus: | 0.204 | Durbin-Watson: | 2.412 |
|---|---|---|---|
| Prob(Omnibus): | 0.903 | Jarque-Bera (JB): | 0.002 |
| Skew: | -0.012 | Prob(JB): | 0.999 |
| Kurtosis: | 3.017 | Cond. No. | 41.6 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
10.2.1.1 R-style formulas
The ability to enter formulas directly into the model is one of the strong sides of statsmodels.formula. This makes it easy to directly specify the dependent and independent variables given we have a data frame, there is no need to create an explicit design matrix (like in sklearn). The way to write the formulas originates from R where they are used in a similar fashion, that’s why they are called “R-style” formulas.
10.2.1.1.1 Specifying dependent and independent variables
The central part of the model formula is dependent - tilde -
independent specification. We can write "y ~ x" and this will be
understood as a linear regression model
\[\begin{equation}
y_i = \beta_0 + x_i \cdot \beta_1 + \epsilon_i.
\end{equation}\]
In particular what is before tilde ~ will be dependent variable
(here \(y\)), and what is after tilde are independent variables (here
just \(x\)). Unless explicitly requested not to, the model also include
the intercept \(\beta_0\).
In case we have more independent variables, those can be added with
plus sign, for instance "y ~ x1 + x2 + x3". This corresponds to the
regression model
\[\begin{equation}
y_i = \beta_0 + x_{i1} \cdot \beta_1 + x_{i2} \cdot \beta_2 + x_{i3} \cdot \beta_3 + \epsilon_i.
\end{equation}\]
Hence the plus sign in the formula does not mean “add” but “include
into the model”. So the formula should be read as “use \(y\) as the
dependent variable, and include \(x_1\), \(x_2\) and \(x_3\) as explanatory
variables.
For instance, if we want to estimate the median house value (medv) as a function of number of rooms (rm), percentage of lower-status population (lstat) and closeness to Charles river (chas), we can run the regression model like:
10.2.1.1.2 Categorical variables and interaction effects
Besides just specifying the variables, the formulas allow many more options. We use titanic data for the examples below.
First, note that if the variable is categorical (i.e. string) by design, it will be automatically converted to approriate dummies. For instance, sex is a categorical variable (with categories female and male) and hence it will be automatically converted into dummies. For instance, if we want to estimate a model \[\begin{equation} \text{survived}_i = \beta_0 + \text{male}_i \cdot \beta_1 + \epsilon_i, \end{equation}\] we can just insert string variable sex into the model:
titanic = pd.read_csv("../data/titanic.csv.bz2")
m = smf.ols("survived ~ sex", data=titanic).fit()
m.summary()| Dep. Variable: | survived | R-squared: | 0.280 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.279 |
| Method: | Least Squares | F-statistic: | 507.1 |
| Date: | Tue, 13 Jan 2026 | Prob (F-statistic): | 3.78e-95 |
| Time: | 17:00:34 | Log-Likelihood: | -697.97 |
| No. Observations: | 1309 | AIC: | 1400. |
| Df Residuals: | 1307 | BIC: | 1410. |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 0.7275 | 0.019 | 38.049 | 0.000 | 0.690 | 0.765 |
| sex[T.male] | -0.5365 | 0.024 | -22.518 | 0.000 | -0.583 | -0.490 |
| Omnibus: | 37.339 | Durbin-Watson: | 1.611 |
|---|---|---|---|
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 39.798 |
| Skew: | 0.419 | Prob(JB): | 2.28e-09 |
| Kurtosis: | 2.834 | Cond. No. | 3.11 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
The line sex[T.male] shows the effect of male dummy with female
being the reference group. This is because the categories will be
ordered alphabetically, and the first (female) taken as the
reference category.
But sometimes the categories are not strings but have numeric lables. For instance, male may be labeled as 1 and female as 2. Or, as is the case of titanic, we have numeric labels for passenger classes. Let us analyze the survival by passenger class using a model like \[\begin{equation} \text{survived}_i = \beta_0 + \text{class2}_i \cdot \beta_1 + \text{class3}_i \cdot \beta_2 + \epsilon_i, \end{equation}\] If we just specify the model as
pclass will be treated as a numeric variable, and the estimate
should be interpreted as the effect per “one unit larger pclass”.
This clearly does not make any sense. Instead, we have to tell
statsmodels that pclass is a categorical variable by wrapping
pclass into C function:
| Dep. Variable: | survived | R-squared: | 0.098 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.096 |
| Method: | Least Squares | F-statistic: | 70.69 |
| Date: | Tue, 13 Jan 2026 | Prob (F-statistic): | 7.10e-30 |
| Time: | 17:00:34 | Log-Likelihood: | -845.26 |
| No. Observations: | 1309 | AIC: | 1697. |
| Df Residuals: | 1306 | BIC: | 1712. |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 0.6192 | 0.026 | 24.084 | 0.000 | 0.569 | 0.670 |
| C(pclass)[T.2] | -0.1896 | 0.038 | -5.011 | 0.000 | -0.264 | -0.115 |
| C(pclass)[T.3] | -0.3639 | 0.031 | -11.732 | 0.000 | -0.425 | -0.303 |
| Omnibus: | 14135.138 | Durbin-Watson: | 1.846 |
|---|---|---|---|
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 142.320 |
| Skew: | 0.446 | Prob(JB): | 1.25e-31 |
| Kurtosis: | 1.653 | Cond. No. | 4.51 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Now pclass will automatically be converted into dummies with 1
being the reference catogory. The lines C(pclass)[T.2] and
C(pclass)[T.3] describe the effect of 2nd and 3rd class travel
respectively (and reveal a sad story of lower survival chance for
lower-class passengers).
We may also want to introduce gender and class interaction effects to test whether the
survival rate depends on class, and whether this dependence differs
for mean and women. This can be achieved with interaction effects in
the form \(\text{male} \times \text{class}\), the full model would look like
\[\begin{align*}
\text{survived}_i &= \beta_0 +
\text{class2}_i \cdot \beta_1 + \text{class3}_i \cdot \beta_2 +
\\
& +
\text{male}_i \cdot \beta_3 +
\\
& +
\text{class2}_i \cdot \text{male}_i \cdot \beta_4 +
\text{class3}_i \cdot \text{male}_i \cdot \beta_5 +
\epsilon_i.
\end{align*}\]
Interaction effects can be intered with asterisk *. For instance,
x1 * x2 will include three variables: \(x_1\), \(x_2\), and \(x_1 \times x_2\). If any of the variables is categorical (either string or
converted to categorical using C, the interaction effects will be
inserted for all categories. Hence the Titanic class and sex example
can be done as:
| Dep. Variable: | survived | R-squared: | 0.370 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.368 |
| Method: | Least Squares | F-statistic: | 153.2 |
| Date: | Tue, 13 Jan 2026 | Prob (F-statistic): | 4.02e-128 |
| Time: | 17:00:34 | Log-Likelihood: | -609.86 |
| No. Observations: | 1309 | AIC: | 1232. |
| Df Residuals: | 1303 | BIC: | 1263. |
| Df Model: | 5 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 0.9653 | 0.032 | 29.974 | 0.000 | 0.902 | 1.028 |
| C(pclass)[T.2] | -0.0785 | 0.049 | -1.587 | 0.113 | -0.176 | 0.019 |
| C(pclass)[T.3] | -0.4745 | 0.042 | -11.414 | 0.000 | -0.556 | -0.393 |
| sex[T.male] | -0.6245 | 0.043 | -14.436 | 0.000 | -0.709 | -0.540 |
| C(pclass)[T.2]:sex[T.male] | -0.1161 | 0.064 | -1.801 | 0.072 | -0.243 | 0.010 |
| C(pclass)[T.3]:sex[T.male] | 0.2859 | 0.054 | 5.340 | 0.000 | 0.181 | 0.391 |
| Omnibus: | 113.251 | Durbin-Watson: | 1.746 |
|---|---|---|---|
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 142.526 |
| Skew: | 0.805 | Prob(JB): | 1.12e-31 |
| Kurtosis: | 3.136 | Cond. No. | 13.3 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
As visible from the table, all the variables from the model above are
included, and the all the pclass categories are taken into account.
The lines containing colon, C(pclass)[T.2]:sex[T.male] and
C(pclass)[T.3]:sex[T.male], are the interaction effects for male
and the corresponding passenger class.
10.2.1.1.3 Transforming variables
## [1] 1000 10One can perform certain computations directly in the model formula. In this example we use a random subset of diamonds data from R’s ggplot2 package. The dataset contains information on diamonds’ price (variable price), mass (carat), and other characteristics we do not discuss here. A few sample lines of the dataset are
## carat cut color clarity depth table price x y z
## 0 0.40 Premium F VVS2 60.7 58.0 1163 4.75 4.70 2.87
## 1 1.01 Very Good H SI1 58.5 56.0 4619 6.60 6.69 3.89
## 2 0.30 Good G VVS1 64.0 57.0 878 4.27 4.23 2.72
## 3 0.71 Ideal E SI1 61.1 57.0 3128 5.75 5.77 3.52
## 4 0.33 Premium H VS1 59.0 62.0 693 4.52 4.49 2.66We model the price as a function of mass. First we use a linear-linear regression model \[\begin{equation} \text{price}_i = \beta_0 + \text{carat}_i \cdot \beta_1 + \epsilon_i: \end{equation}\]
| Dep. Variable: | price | R-squared: | 0.845 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.845 |
| Method: | Least Squares | F-statistic: | 5452. |
| Date: | Tue, 13 Jan 2026 | Prob (F-statistic): | 0.00 |
| Time: | 17:00:35 | Log-Likelihood: | -8808.8 |
| No. Observations: | 1000 | AIC: | 1.762e+04 |
| Df Residuals: | 998 | BIC: | 1.763e+04 |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -2367.8150 | 100.417 | -23.580 | 0.000 | -2564.869 | -2170.761 |
| carat | 7993.6184 | 108.256 | 73.840 | 0.000 | 7781.184 | 8206.053 |
| Omnibus: | 200.672 | Durbin-Watson: | 1.935 |
|---|---|---|---|
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 2033.544 |
| Skew: | 0.602 | Prob(JB): | 0.00 |
| Kurtosis: | 9.881 | Cond. No. | 3.66 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
The results reveal that the price is closely related to the carat weight (coefficient value 7993.6 with a huge \(t\)-value). The model is also fairly good in terms of \(R^2 = 0.845\). But the plot below reveals that linear-linear model is not the best fit for the data:
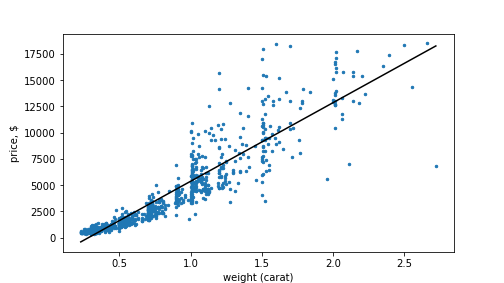
plot of chunk diamondPlot
Diamonds’ price as a function of weight. Linear-linear scale.
The figure suggest that the true price-weight curve is more convex,
not the fact that the low-weight diamonds’ (ct < 0.5) price is consistently
underpredicted, while those in range 0.5-1 are overpredicted.
We may try log-log transformation instead. We may use functions
directly to transform variables, in this case we can use np.log for
log transform and write np.log(price) instead of price:
\[\begin{equation}
\log\text{price}_i = \beta_0 + \log\text{carat}_i \cdot \beta_1 + \epsilon_i:
\end{equation}\]
| Dep. Variable: | np.log(price) | R-squared: | 0.932 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.932 |
| Method: | Least Squares | F-statistic: | 1.362e+04 |
| Date: | Tue, 13 Jan 2026 | Prob (F-statistic): | 0.00 |
| Time: | 17:00:35 | Log-Likelihood: | -123.32 |
| No. Observations: | 1000 | AIC: | 250.6 |
| Df Residuals: | 998 | BIC: | 260.4 |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 8.4579 | 0.010 | 809.901 | 0.000 | 8.437 | 8.478 |
| np.log(carat) | 1.7035 | 0.015 | 116.714 | 0.000 | 1.675 | 1.732 |
| Omnibus: | 17.992 | Durbin-Watson: | 1.912 |
|---|---|---|---|
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 33.278 |
| Skew: | -0.005 | Prob(JB): | 5.94e-08 |
| Kurtosis: | 3.894 | Cond. No. | 2.06 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
This model looks better as suggested by both \(R^2 = 0.932\) and the \(t\)-value. And here is the corresponding plot:
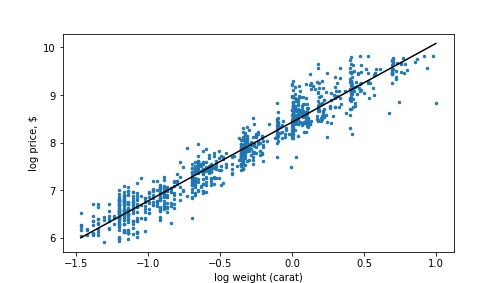
plot of chunk diamonds-log-log
Diamonds’ price as a function of weight. Log-log scale.
Now the trend line looks perfectly straight and the linear model fits very well.
10.2.2 Scikit-learn and LinearRegression
Another very popular python library for linear regression is scikit-learn (usually abbreviated as sklearn). While statsmodels is mostly geared toward inference and provides nice summary tables with coefficients, \(t\) statistics and \(R^2\); sklearn is designed for predictive modeling. It does not provide similar output tables, and it does not have formula API. Instead, one has to create the design matrix manually, and use this when fitting the model. This may be somewhat complex if one needs some feature engineering, such as creating dummies. As an advantage though, it contains a large number of other predictive models with similar API, so swapping one model for another is very easy. It also contains other tools, such as cross-validation, that are commonly used for predictive modeling. However, some of the methods are implemented slightly differently than the textbook cases in statsmodels, in particular sklearn-versions often contain regularization switched on by default. So the results may turn out to be slightly different than what one may expect based on other implementations.
Below, we demonstrate how to use sklearn’s LinearRegression, Section 11.2.2 below explains how to perform logistic regression.
10.2.2.1 Numeric variables only
Let us start with the example of estimating the petal width of
versicolor flowers (see Section 10.1.1).
Linear regression can be
performed using LinearRegression function:
# we have to import the model..
from sklearn.linear_model import LinearRegression
# .. and create the model
m = LinearRegression()The import call makes the LinearRegression function available. The
second line of code constructs
the linear regression object that we can fit afterwards. It is
similar to statsmodels call m = smf.ols("y ~ x", data=df),
i.e. setting up the model without fitting it.
LinearRegression takes
various parameters, but here we are usually
happy with the defaults. In particular, we are happy with the dafault
behavior of adding the intercept to the model automatically, so we
do not have to add a constant column to \(\mathsf{X}\).
Next, we have to construct the design matrix. Here it is easy, as it just contains a single numeric variable. But it must be a matrix (or data frame), not a vector. The following code fill fail:
## create design matrix
X = versicolor.plength
# problem: X is a series, not matrix!
m = LinearRegression()
_t = m.fit(X, versicolor.pwidth)## ValueError: Expected a 2-dimensional container but got <class 'pandas.core.series.Series'> instead. Pass a DataFrame containing a single row (i.e. single sample) or a single column (i.e. single feature) instead.Let us now ensure that our design matrix is a matrix and not a
vector. We can achieve this by adding additional brackets around
pl (see Section 3.3.1 for more explanations):
# create design matrix
X = versicolor[["plength"]]
# now X is a data frame
m = LinearRegression().fit(X, versicolor.pwidth)Note the similarities and differences between statsmodels and
sklearn: in both cases you first set up the model (either with
ols() or LinearRegression()) and thereafter fit it with .fit()
method. However, in case of statsmodels you supply data to the
ols() function, in case of sklearn you supply it to .fit()
method. Also, sklearn requires you to supply the design matrix X
and target y separately. statsmodels will create these objects
itself from the formula.
Now m is a fitted model. We can query the parameters as
## -0.08428835489833553## array([0.3310536])You can see that these numbers are exactly the same as in Section @ref(#linear-regression-statsmodels).
One can query a quick model goodness summary using .score method.
In case of linear regression (and other regression models) it returns
\(R^2\):
## 0.6188466815001423Again, we get exactly the same number as when using statsmodels
above. Note that .score method expects two arguments: the design
matrix X and target y, in this order. (It is the same order as
for the
.fit method.) Internally it predicts the values for
every row of X, computes errors between predictions and y, and
transforms this to \(R^2\). But by supplying X and y to the
.score-method we can calculate \(R^2\) on different data–on
validation data instead of training data (see Section
13 for more information).
But sklearn does not have a function to print a similar summary
table as what .summary does in
statsmodels. We cannot easily see standard errors, \(p\)-values or
statistical significance.
sklearn is designed for predictive modeling where such tables are of
little value. In complex
cases, for instance in image processing, we may want to include values
of millions of
pixels into the model and here individual specification of each pixel
is clearly infeasible. We prefer to use the design matrix approach
instead. In a similar fashion, we are not interested in interpreting
values of
millions of coefficients. sklearn is designed for exactly such
tasks.
TBD: exercise with a single variable (Hubble)
Exercise 10.2 Take the iris data. Extract data for virginica (where Species = virginica) and run a regression: \[\begin{equation} \text{sepal width}_i = \beta_0 + \beta_1 \cdot \text{sepal length}_i + \beta_2 \cdot \text{petal width}_i + \beta_3 \cdot \text{petal length}_i + \epsilon_i. \end{equation}\] Compute \(R^2\).
See the solution
10.2.2.2 Categorical variables
As sklearn does not include a formula method, one cannot rely on it
to convert categoricals to
dummies. Instead, one has to make dummies manually. A
good choice will be to use pd.get_dummies
function. As an example,
let’s use the males
dataset
and estimate a regression model
\[\begin{equation}
\log w_i = \beta_0 + \beta_1 \cdot \text{school}_i +
\beta_2 \cdot \text{residence}_i +
\epsilon_i.
\end{equation}\]
Here \(\log w\) is log hourly wage (in 1980-s, called wage in the dataset),
school is years of
schooling, and residence is the residence region (“rural_area”,
“north_east”, “nothern_central” and “south”).
As the first step, we have to create the design matrix X by including all the relevant variables and converting categoricals to dummies:
males = pd.read_csv("../data/males.csv.bz2", sep="\t")\
[["wage", "school", "residence"]]
# a look at the original data
males.sample(3)## wage school residence
## 673 1.121050 10 south
## 1631 1.933081 12 nothern_central
## 701 1.072815 12 southThe simplest approach we can do is just to convert all the explanatory
variables (here school and residence) with pd.get_dummies and
let it sort out what is numeric and what is not. We also
ask one category to be dropped as the reference category:
X = pd.get_dummies(males[["school", "residence"]],
# do not include 'wage' into X!
drop_first=True)
X.sample(3)## school ... residence_south
## 4071 9 ... False
## 1587 15 ... False
## 2323 12 ... False
##
## [3 rows x 4 columns]One can see that school was preserved as a numeric column, but the four-category residence was transformed into three columns (with north_east dropped as the reference category).
For consistency, we also separate the outcome, \(\log w\), into a vector \(\mathbf{y}\):
Now this is a ready-made numeric matrix we can use for fitting
sklearn models. We also need the outcome variable, here just the
log-income males.wage. Now we can fit the model as
we assign the result of .fit to a temporary variable—it returns
the fitted object and we do not want it to be printed here.
As no output is visible, we may manually examine the results:
## array([ 0.07916899, -0.08389827, -0.14039362, -0.08565235])We can see that .coef_ is a vector of four values–one for each
column of X, in the same order as in the columns. So .coef[0]
tells us that those who have spent one year more at school earn 8%
more, at least in average. We also see that income in all regions is
smaller than in the reference region, North-East.
We can compute \(R^2\) using the .score method, there are no
differences with the only numeric case in Section
10.2.2.1:
## 0.07031991903276569Exercise 10.3 Take the males dataset and run a regression: \[\begin{equation} \log w_i = \beta_0 + \beta_1 \cdot \text{school}_i + \beta_2 \cdot \text{ethn}_i + \epsilon_i. \end{equation}\] where school is years of schooling, and ethn is race categories.
Ensure you have correct number of coefficients. Compute \(R^2\).
See the solution
10.3 Model Goodness
10.3.1 Create Random Data for Experiments
Random data is useful for learning or testing methods for various reasons as they allow to design exactly the features we are interested in trying our methods on, and when manually creating data, we will also know what exactly do we put in there and hence also what we ought to get out. If our results do not agree, something is wrong either in our code or our understanding of the method.
For linear regression we can create data exactly as specified by the linear regression model: \[\begin{equation} y_i = \beta_0 + \beta_1 \cdot x_i + \epsilon_i. \end{equation}\] Here \(x\) is our explanatory variable. We do not have to specify precisely how it looks like, but random normals are a good bet. \(\epsilon\) is the random disturbance term, a term necessary to knock the points off from the straight line to make the artificial data to resemble real data. This should be normal to ensure that the estimated standard errors are correct. We also have to choose the values for parameters \(\beta_0\) and \(\beta_1\), in this example we choose 1 and 2. Let us create a dataset with 100 observations:
n = 100
beta0, beta1 = 1, 2
x = np.random.normal(size=n) # create random x
eps = np.random.normal(size=n) # random disturbance term
y = beta0 + beta1*x + eps # create y exactly as specified by linear regression definiton
# finally, keep this data in a dataframe for future
randomDF = pd.DataFrame({"x":x, "y":y})Let’s plot the resulting \(x\) and \(y\):
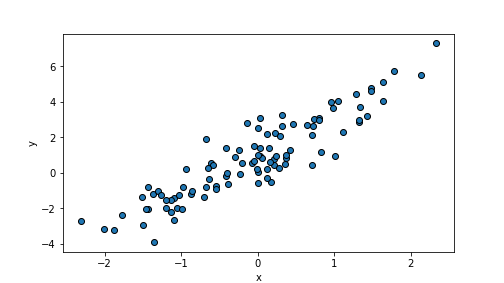
plot of chunk unnamed-chunk-35
We can see an upward-trending cloud of points.
Exercise 10.4
- Modify the parameters \(\beta_0\) and \(\beta_1\) and create a downward trending point cloud
- Modify the
scaleparameters ofnp.random.normalwhen creating \(\epsilon\) to create a point cloud that is very narrow (dots are almost perfectly lined up).
Show your data on a plot.
See the solution
10.3.2 Compute SSE, TSS, and \(R^2\) manually
In this example we use our random data frame we created above. As the first step, let’s compute TSS using the definition \[\begin{equation} \text{TSS} = \sum_{i=1}^{N} (y_{i} - \bar{y} )^{2}. \end{equation}\] This definition can be directly translated into computer code:
## 499.9520198054768This is the total variation in the data (in \(y\)). How much will be left unexplained by linear regression? First we have to fit a linear regression model in order to be able to compute this:
m = smf.ols("y ~ x", data=randomDF).fit() # fit the model
beta0 = m.params[0] # assign the best solution coefficients to beta0, beta1## <string>:1: FutureWarning: Series.__getitem__ treating keys as positions is deprecated. In a future version, integer keys will always be treated as labels (consistent with DataFrame behavior). To access a value by position, use `ser.iloc[pos]`Now we can compute the predicted values in a fairly similar way as when we created random data. Just remember that we compute expected values as predictions, and hence we just ignore \(\epsilon\):
Now we have both true values y and predicted values yhat. We can
use the definition of SSE and compute deviations \(e\) and thereafter
SSE:
## 86.00291587805127Finally, we can also compute \(R^2\) from the definition:
## 0.827977660913314We may want to compare our “home-made” \(R^2\) with the one provided by
statsmodels rsquared attribute:
## 0.827977660913314One can see that the the summary method provide a similar \(R^2\).
Exercise 10.5 Use the versicolor data to estimate a regression model \[\begin{equation} \text{Petal width}_{i} = \beta_{0} + \beta_{1} \cdot \text{Petal length} + \epsilon_{i}, \end{equation}\] the exact same model we used in Solving Linear Regression Tasks Manually. Compute \(R^2\) manually as we did above.
See the solution
Exercise 10.6 Modify your data generation procedure, in particular the scale
parameter for \(\epsilon\) so that you get:
- \(R^2 > 0.99\)
- \(R^2 < 0.1\).
Show the corresponding data points on a plot.
See the solution