Chapter 11 Doing statistics with R
11.1 Random numbers
R has a very handy built-in random number functionality. It covers a
number of common distributions, and allows to generate random numbers
and calculate probability density (or mass), cumulative distribution,
and distribution quantiles. All these functions, except sample(),
use a standard
interface.
11.1.1 Sampling elements: sample()
sample() samples elements from a vector. Its basic usage is
sample(vector, n) that pulls out \(n\) random elements from the
vector. For instance,
sample(1:10, 3)## [1] 5 6 3This takes three random elements out of the vector 1:10, i.e. out of
numbers 1 to 10. By default, it extracts the elements without
replacement, i.e. if number 5 is already taken, you cannot get
that number again. If this is not what you wish, you can specify
replace = TRUE to choose elements with replacement:
sample(c("A", "B"), 10, replace = TRUE)## [1] "B" "A" "A" "B" "B" "A" "B" "B" "A" "B"This selects either “A” or “B” randomly for 10 times. This is obvioulsy impossible unless with select with replacement.
There is a handy shortcut to select numbers 1, 2, …, K: sample(K, n). So in order to emulate rolling a die you can write
sample(6, 1) # one number between 1 and 6## [1] 5Exercise 11.1 You want to simulate rolling 5 dice. You write
sample(6, 5)Is this correct?
One can also ask sample() to do non-uniform sampling. For instance
sample(c("A", "B"), 10, replace = TRUE,
prob = c(3, 7))## [1] "B" "B" "A" "B" "B" "A" "B" "B" "B" "B"will result in “A” sampled with 30% and “B” sampled with 70% probability.
sample() is a somewhat special function among random number
generation, as it does not follow the standard API.
11.1.2 Replicable random numbers
Random numbers are, well, random. This means each time you generate those, you’ll get a different values:
sample(6, 3) # one set of values## [1] 5 2 3sample(6, 3) # different values## [1] 2 4 6This is often all you need. But sometimes you want your random numbers to be replicable. This can be acheived by setting the “seed”–the initial values of the random number generators.
set.seed(4)
sample(6, 3) # one set of values## [1] 3 6 5set.seed(4)
sample(6, 3) # the same set of values## [1] 3 6 5set.seed() applies to all random numbers, not just those generated
with sample().
11.1.3 r..., d..., p..., q...: random numbers and distributions
Random numbers in R are related to probability distribution. Each
distribution has its short name (e.g. norm for normal), and four
functions that provide the related functionality:
r...(n)(e.g.rnorm(3)) to sample \(n\) random numbers of this distribution.d...(x)(e.g.dnorm(0)) to compute probability density (or probability mass) at \(x\)p...(x)(e.g.pnorm(0)) to compute the cumulative probabilityq...(x)(e.g.qnorm(0.5)) to compute the distribution quantiles
These functions typically also take additional arguments, such as location, scale, and shape parameters.
Let’s demonstrate the usage with normal distribution as an example.
The short name of the normal functions is norm, i.e. the functions
are called rnorm(), dnorm(), pnorm() and qnorm().
We can create random normals with rnorm():
rnorm(4)## [1] 0.8911446 0.5959806 1.6356180 0.6892754Or if we want to specify a different mean and standard deviation:
rnorm(5, mean = 100, sd = 10)## [1] 87.18753 97.86855 118.96540 117.76863 105.66604The normal density can be computed as
dnorm(0)## [1] 0.3989423dnorm(0, mean = 100, sd = 10)## [1] 7.694599e-24And the cumulative density as
pnorm(0.5)## [1] 0.6914625You can use the base-R function curve() to visualize the pdf-s and
cdf-s:
curve(dnorm, -5, 5, ylim = c(0, 1))
curve(dnorm(x, mean = 3, sd = 0.5),
col = "red", add = TRUE)
curve(pnorm(x, mean = 3, sd = 0.5),
col = "green", lty = 2, add = TRUE)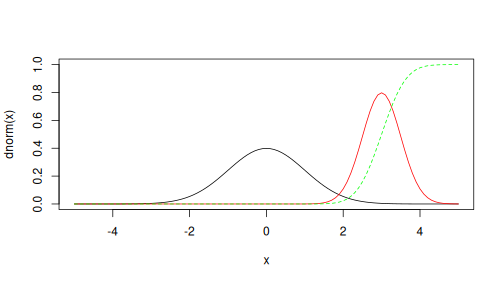
norm for normal distribution, the other distribution include
binomfor binomial (and Bernoulli). It needs two parameters, sample size and probabilitylnormfor log-normalpoisfor Poisson. It requires one parameter, the expected value.expfor exponentialgammafor gamma distribution. It requires a shape parameter.betafor beta distribution. It requires two shape parameters.
There are more built-in distribution, and add-on packages provide even more distributions.
11.2 Simulating statistical models
Random numbers are extremely useful when you want to test and try out statistical models.
11.2.1 A simple case: law of large numbers
For instance, let’s demonstrate the Law of Large Numbers–the
sample average is approaching the expected value as the sample size
\(N\to\infty\). In order to demonstrate this, we need to pick a random
process, average of which we are analyzing. Let’s use
a very simple one–flip two coins and compute
the numbre of heads. Two coins can be simulated as rbinom(2, 1, 0.5), we’ll replicate this many times, and compute the average.
If the number of replications \(R\)
is large, the
average number of heads should be close to unity.
- create a vector to store the number of heads in each replication
- for each experiment do the following:
- flip two fair coins using
rbinom(2, 1, 0.5). This will result in two numbers, both either “0” or “1”. - compute the total number of heads (just sum these numbers)
- save the sum into the vector
- flip two fair coins using
- compute the average of the number of heads that are stored in the vector.
Thereafter we can repeat the above for different number of experiments.
Let’s do the code for 10 experiments:
S <- 2 # how many coins do you flip
R <- 10 # how many experiments
heads <- numeric(R) # number of heads in each experiment
for(i in 1:R) {
coins <- rbinom(S, 1, 0.5)
heads[i] <- sum(coins)
}
heads # show the results of every experiement## [1] 1 2 1 2 1 2 1 1 0 1You can see that we’ll get mostly either 1 or 2 heads, but in one case also no heads. Finally, we are not interested in every single result but the average number of heads over experiments:
mean(heads)## [1] 1.2Note that these simulations involve two sample size: one, denoted by \(S\) is the number of coins, and the other, denoted by \(R\), is the number of replications–how many time do we flip \(S\) coins.
This was a single experiment with 10 replications. If we want to see
how LLN works, we need to repeat it
for different number of replications, e.g. from 1 to 1000, in a loop.
Each time we compute
the average number of heads, and we need a vector
to store those averages (called avgs below):
N <- 1000 # the max # of experiments
avgs <- numeric(N) # experiment averages
for(R in 1:N) {
heads <- numeric(R) # number of heads in each experiment
for(i in 1:R) {
coins <- rbinom(2, 1, 0.5)
heads[i] <- sum(coins)
}
avgs[i] <- mean(heads)
}
head(avgs) # just check -- take a look at a few values ## [1] 1.000000 1.500000 1.666667 1.000000 1.400000 1.000000Now avgs contains the average number of heads for 1, 2, … \(R\)
trials.
Finally, let’s make a plot to show how the average depends on \(R\):
plot(avgs, xlab = "R", ylab = "Average",
type = "l")
axis(4) # add axis on right side of the plot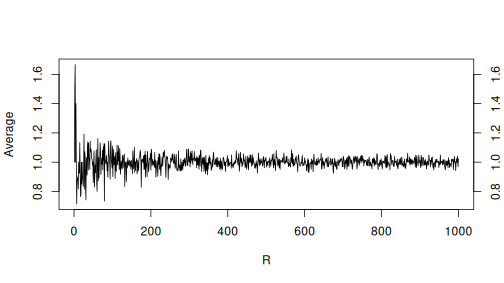
The plot shows how the initial large wobbles at small \(R\) values converge to close to one–the expected value.
11.2.2 Expected value and sample average
Computer simulations is an excellent way to check the properties of RV-s. For instance, consider a RV: \[\begin{equation*} Y = \begin{cases} 0 & \text{with probability}\quad 0.5\\ 1 & \phantom{\text{with probability}}\quad 0.25\\ 2 & \phantom{\text{with probability}}\quad 0.25.\\ \end{cases} \end{equation*}\] It is easy to show that \(\mathbb{E} X = 0.75\) and its variance \(\mathrm{Var} X = 0.6875\) (see Lecture Notes).
You can sample from an arbitrary RV using the sample() function by
providing the respective values and probabilities (see Section
11.1.1). For this RV, you can write
sample(c(0, 1, 2), 10, prob = c(0.5, 0.25, 0.25),
replace = TRUE)## [1] 0 0 1 0 1 1 2 0 0 0Now you should just create a large sample, and compute its mean and variance:
N <- 10000
x <- sample(c(0, 1, 2), N, prob = c(0.5, 0.25, 0.25),
replace = TRUE)
mean(x)## [1] 0.752var(x)## [1] 0.6909651The results are indeed quite close to the theoretical values. It is also very easy to simulate functions of this RV (see Lecture Notes). For instance, \(\mathbb{E}X\) can be simulated as
mean(x^2)## [1] 1.2564It is close to the true value \(1.25\).
Exercise 11.2
Consider a RV \[\begin{equation*} Y = \begin{cases} -10 & \text{with probability}\quad 0.1\\ 0 & \text{with probability}\quad 0.5\\ 1 & \phantom{\text{with probability}}\quad 0.4.\\ \end{cases} \end{equation*}\] First compute and then simulate- \(\mathbb{E}X\)
- \(\mathrm{Var}X\)
- \(\mathbb{E} \mathrm{e}^X\)
- \(\mathrm{Var} \mathrm{e}^X\)
TBD: exercise: sample quantiles
11.2.3 Linear regression
Linear regression has a number of assumptions, one of which is that the error term is normally distributed. If the model is \[\begin{equation} y_i = \beta_0 + \beta_1 \cdot x_i + \epsilon_i \end{equation}\] then \[\begin{equation} \epsilon_i \sim N(0, \sigma^2). \end{equation}\]
Let’s make the following experiment: assume there is no relationship between \(x\) and \(y\). We sample data of size \(N\). How often do we see a relationship just because of the sample is random?
First we need to decide about the parameters \(\beta_0\) and \(\beta_1\). Assuming there is no relationship is equivalent to setting \(\beta_1 = 0\). The assumption does not say anything about \(\beta_0\) though–it is essentially average \(y\) value and we can pick any number. For simplicity, we can pick \(\beta_0 = 0\) as well.
Next, we need to specify \(x\) values. As these are not stochastic, we
can, again, pick all sorts of numbers, such as 1, 2, …, 10; or just
random numbers here too. Let’s imagine our data only contains numbers
1 to 10, so we can randomly sample from these as sample(10, N, replace = TRUE).
The most important step is to model the relationship between \(y\) and \(x\). As we assume there no relationship, \(y\) is just equal to the error term \(\epsilon\). But below we still use the full specification \[\begin{equation} y = \beta_0 + \beta_1 \cdot x + \epsilon \end{equation}\] for consistency, using \(\beta_0 = \beta_1 = 0\).
Assume the standard assumption is correct and the error term follows the standard normal \(\epsilon \sim N(0, 1)\). (We can also see what happens if this assumption is violated, but let’s leave this exercise for another time.)
Finally, let’s look at a small sample of size \(N = 10\). So a single dataset can be generated as
beta0 <- beta1 <- 0
N <- 10
x <- sample(10, N, replace = TRUE)
eps <- rnorm(N)
y <- beta0 + beta1*x + eps
plot(x, y)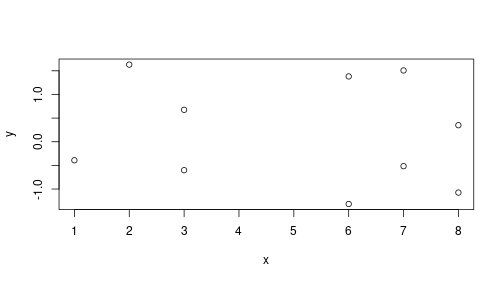
As expected, if \(\beta_1 = 0\), we do not see much of a relationship between \(x\) and \(y\). This can be confirmed with linear regression:
m <- lm(y ~ x)
summary(m)##
## Call:
## lm(formula = y ~ x)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.42482 -0.87463 -0.09974 1.05002 1.46088
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.47935 0.83527 0.574 0.582
## x -0.06186 0.14743 -0.420 0.686
##
## Residual standard error: 1.15 on 8 degrees of freedom
## Multiple R-squared: 0.02154, Adjusted R-squared: -0.1008
## F-statistic: 0.1761 on 1 and 8 DF, p-value: 0.6858\(\beta_1\), denoted here by x, is
-0.062
But how often can we see a relationship, just because of the random nature of sampling? To answer this question, we can repeat the experiment many times, each time estimate a linear regression model, and save the estimated \(\beta_1\). What are the largest values we see there?
We can use a very similar code to what we did above. Generate data
using the example above; fit a linear regression model; extract the
coefficient "x", and save it in a vector:
R <- 1000
beta0 <- beta1 <- 0
N <- 10
betas <- numeric(R)
for(r in 1:R) {
x <- sample(10, N, replace = TRUE)
eps <- rnorm(N)
y <- beta0 + beta1*x + eps
m <- lm(y ~ x)
betas[r] <- coef(m)["x"]
}
hist(betas, breaks = 30)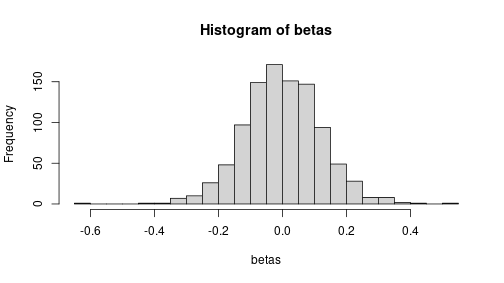 But the largest estimated values are above 0.5 (in absolute value):
But the largest estimated values are above 0.5 (in absolute value):
range(betas)## [1] -0.6447306 0.5119822Even if there is no true relationship, we’ll occasionally see quite a substantial estimates–just because of random sampling.
The standard deviation of the estimated betas is very close to what we computed above:
sd(betas)## [1] 0.1218043This ensures that the statistics, as compute by summary() are
reliable.
Exercise 11.3 The simulation above assumes that the intercept \(\beta_0 = 0\). But what happens if you choose a different \(\beta_0\), e.g. \(\beta_0 = 10\)?
- create a single sample of data, and estimate the linear regression model as above. What is the estimated \(\beta_1\)? How big is its standard error?
- Repeat the above 1000 times and display the histogram of estimated \(\beta_1\), and its standard deviation. Do you get numbers that are close to those in the regression summary table?
- Explain what you find!
Exercise 11.4 The reason that your simulated standard errors are so close to the theoretical ones as provided by the summary table, is that we have fulfilled all the assumptions behind the theoretical standard errors. One of these is that the error term is normally distributed, \(\epsilon \sim N(0, \sigma^2)\). But what happens if the error term is not normally distributed?
- Use three different distributions for the error terms:
- rectangular, standard uniform (\(\epsilon \sim Unif(0,1)\)). You
can sample it as
runif(N). - Asymmetric, exponential (\(\epsilon \sim Exp(1)\)), can be
simulated as
rexp(N, 1) - Very unequal, log-normal (\(\epsilon \sim LN(0, 1.5)\)), can be
simulated as
rlnorm(N, 0, 1.5)
- rectangular, standard uniform (\(\epsilon \sim Unif(0,1)\)). You
can sample it as
- replicate what we did above.
- do the simulated results and theoretical results align?
See the solution