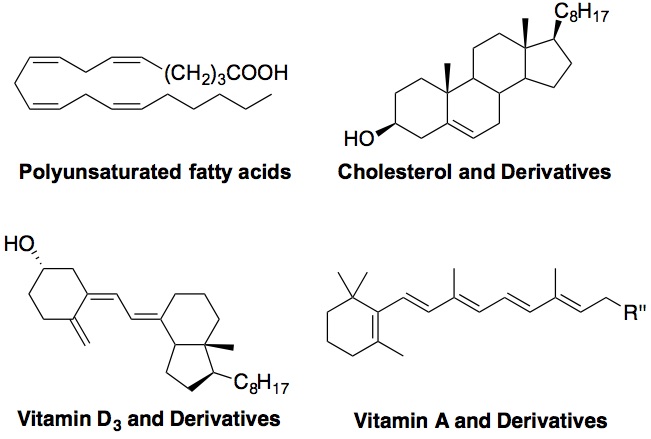
Chemistry of Lipid Peroxidation
Free radical oxidation of polyunsaturated fatty acids (PUFAs) and sterols, i.e., lipid peroxidation, plays important roles in a number of human diseases, such as atherosclerosis, asthma, Alzheimer’s disease, cancer, diabetes, etc. In this project, we aim to 1) develop chemical methods to measure oxidation rate constants of different classes of lipids, including PUFAs, sterols, as well as highly conjugated lipids such as vitamin D3 and carotenoids; and 2) to elucidate the oxidation mechanism of these lipids and identify their products. The ultimate goal is to identify particularly oxidizable biomolecules and to understand their normal and pathological functions in human. Figure below shows oxidizable lipids that are of our interest. Click here for more information.
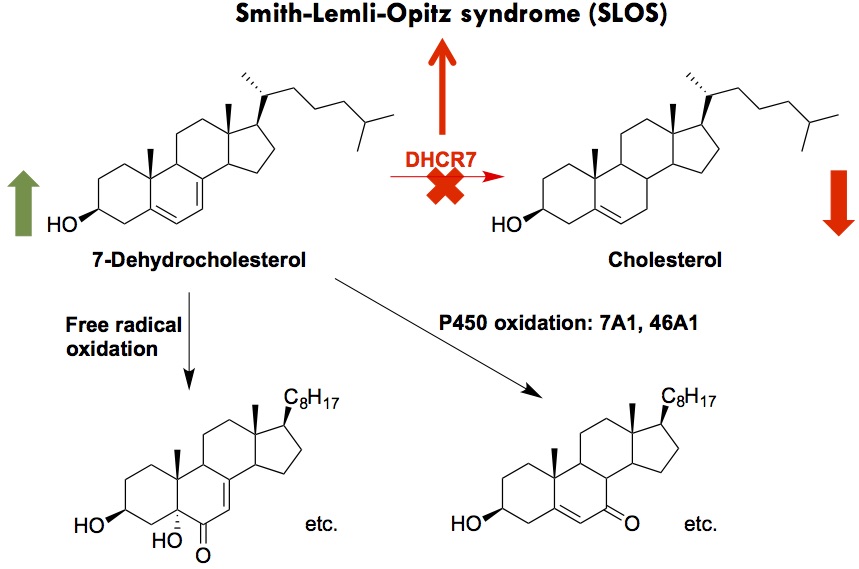
Smith-Lemli-Opitz syndrome (SLOS)
SLOS is a developmental disorder caused by mutations in the gene (DHCR7) encoding 3β-hydroxysterol-Δ7-reductase, the enzyme that catalyzes the reduction of 7-dehydrocholesterol (7-DHC) to cholesterol in the last step of cholesterol biosynthesis [1]. SLOS is characterized by multiple congenital malformations, neurological defects, and autism-like behavior. The carrier frequency of SLOS was found to be 1 in 30 in the US population, predicting an incidence of 1 in 1,590 to 13,500. The genetic defect in SLOS leads to decreased levels of cholesterol and significantly elevated levels of 7-DHC in affected individual.
The cholesterol precursor, 7-DHC, was found to be exceptionally reactive toward free radical oxidation, leading to the formation of over a dozen biologically active oxysterols [2,3]. 7-DHC was also found to be a novel substrate of cytochrome P450s (CYP), leading to the known toxic oxysterol, 7-ketocholesterol, via CYP 7A1, and 24-OH- and 25-OH-7-DHC via 46A1 [4,5]. The goal of this project is to understand the role of 7-DHC-derived oxysterols in the pathophysiology of SLOS and develop therapies that target 7-DHC and/or its oxysterols. We aim to examine the effects of these oxysterols on metabolome and lipidome using mass spectrometry and on transcriptome using qPCR and sequencing. Click here fore more information.
Environmental Toxicology
Drugs and their metabolites, as well as drug-like environmental agents, could display broad and sometimes potent off-target effects, interference with lipid metabolic pathways being one of them. Using in silico structural similarity screening and in vitro assay validation, we found a class of environmental chemicals, benzalkonium chlorides (BACs), as potent inhibitor of DHCR7 (Herron et al., Tox. Sci., 2016). Furthermore, using a multi-omics approach by combining lipidomcs, targeted sterolomics, and RNA sequencing, we found that BACs indeed alter sterol and lipid homeostasis in embryonic brain (Herron et al., Tox. Sci., 2019).
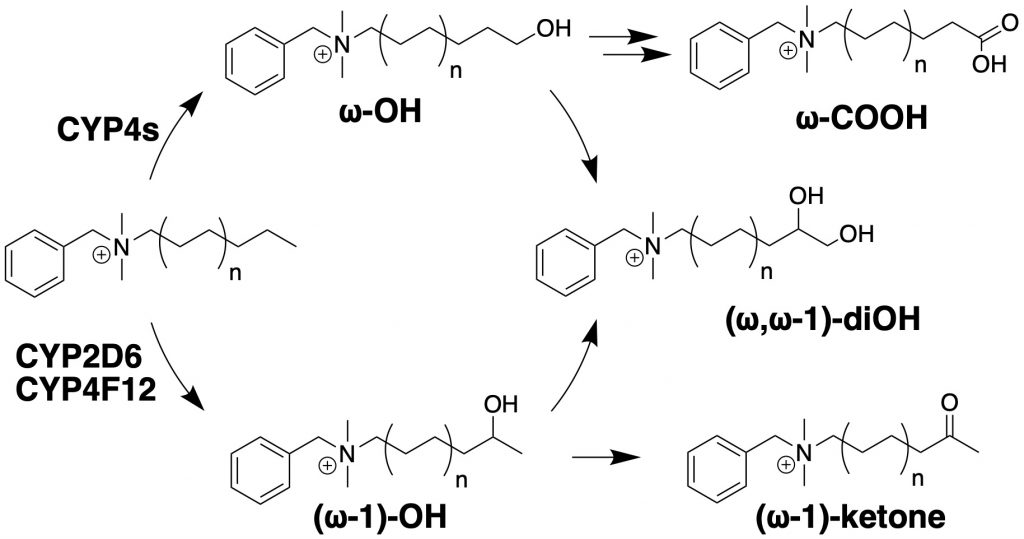
In parallel, we have identified major metabolic pathways of BACs by cytochromes P450 found in human liver, which include omega- and (omega-1)-hydroxylation reactions and subsequent secondary metabolites (Seguin et al., Chem. Res. Tox. 2019). Our ongoing effort include further elucidation of the BAC metabolic pathways and the mechanisms of transport of BACs by transporters using in vitro models, the interactions between metabolism, transport, and toxicity, and the impact of BACs on gut microbiome and gut-liver interactions in animal models and in humans. Check out our recent papers in these areas:
QACs and their metabolites database: Nguyen et al. 2024
Imapct of BACs on gut microbiome and bile acid metabolism: Lopez et al. 2024.
Lipidomics of Antibiotic-Resistant Bacteria
In collaboration with Prof. Brian Werth in the Department of Pharmacy, we are interested in lipidomic and metabolic alterations resulting from development of antibiotic resistance in Gram-positive bacterial pathogens, such as Staphylococcus aureus. Some of the effective drugs against methicillin-resistant S. aureus (MRSA) are cell wall or cell membrane-targeting antimicrobials, such as vancomycin, daptomycin, and dalbavancin. However, resistance to these antimicrobials has emerged. We hypothesize that genetic mutations in the resistant strains would lead to characteristic lipidomic and metabolic changes in the bacteria due to the close interactions between cell wall and cell membrane metabolic processes. We use lipidomics and molecular biology methodologies to investigate these resistance mechanisms. See our publications on lipidomic changes associated with daptomycin resistance (Hines et al. mSphere, 2017), cross-resistance among vancomycin, daptomycin, and dalbavacin, and seesaw effect and synergy with beta-lactams (Hines et al. J. Antimicrob. Chemother. 2020; Zhang et al., Front. Mol. Biosci. 2021).
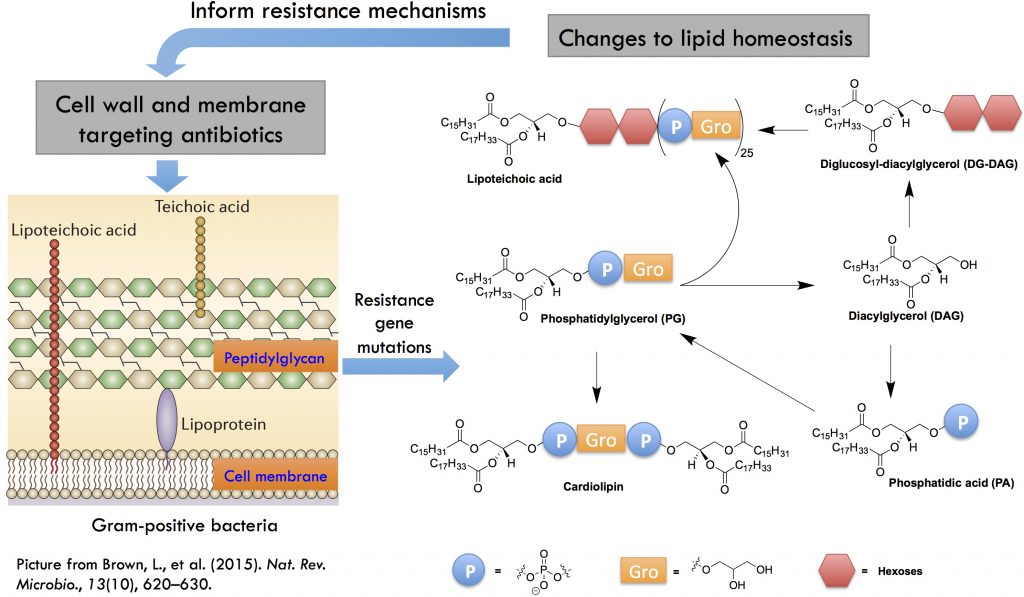
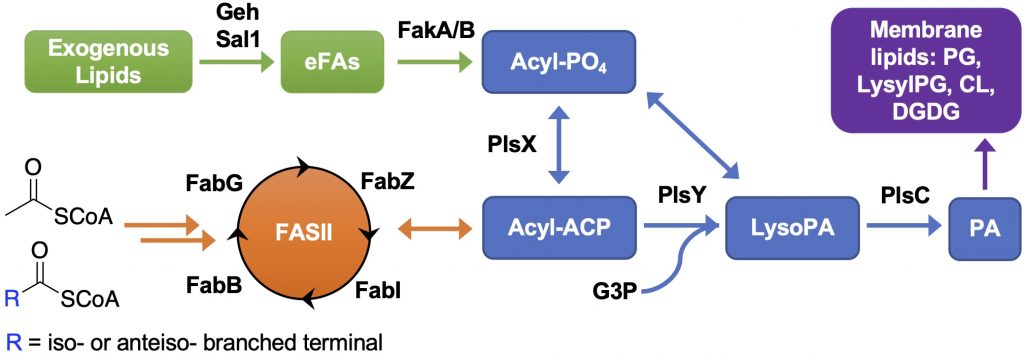
Metabolic Interactions Between the Host and Pathogens
We aim to elucidate the mechanisms underlying the metabolic interactions between host and pathogens and the biological consequences of such interactions. S. aureus utilizes various strategies to survive in the host environment, one of which is to hydrolyze host-derived lipids using secreted lipases and incorporate the hydrolyzed fatty acids into its membrane lipids via fatty acid kinases (FakA). The fatty acid composition in the host and S. aureus are dramatically different, i.e., mammalian cells are composed of straight-chain saturated and unsaturated fatty acids while S. aureus can only synthesize saturated straight-chain or amino acid-derived branched-chain fatty acids via type II fatty acid synthesis (FASII). Furthermore, S. aureus can synthesize PGs, lysyl-PGs, DGDGs, and cardiolipins, but cannot synthesize major mammalian lipids, such as phosphatidylethanolamines (PEs), phosphatidylcholines (PCs), and sphingomyelins (SMs). Through collaboration with Prof. Brian Wilkinson at Illinois State University and Prof. Francis Alonzo at Loyola University Chicago, we found that host serum lipids are hydrolyzed by secreted lipases and then incorporated into S. aureus lipids and that cell wall thickness is increased in the presence of serum. We are interested in 1) How does the incorporation of host fatty acids affect the growth of S. aureus in a host environment and their susceptibility to antimicrobials? and 2) Does the release of host fatty acids by S. aureus lipases detrimentally affect the host cells? Please check out our relevant papers (Hines et al. mSphere 2020; Pruitt et al. mSphere 2023).
Mass Spectrometry
We use liquid chromatography-mass spectrometry (LC-MS) to carry out untargeted and targeted analysis of lipids, drugs, small molecule metabolites, and peptides. We are specialized in lipid analysis, including targeted analysis of sterols and oxysterols and comprehensive lipidomics. Reverse phase and hydrophilic interaction liquid chromatography are used to separate lipid classes, as well as individual species within each class.
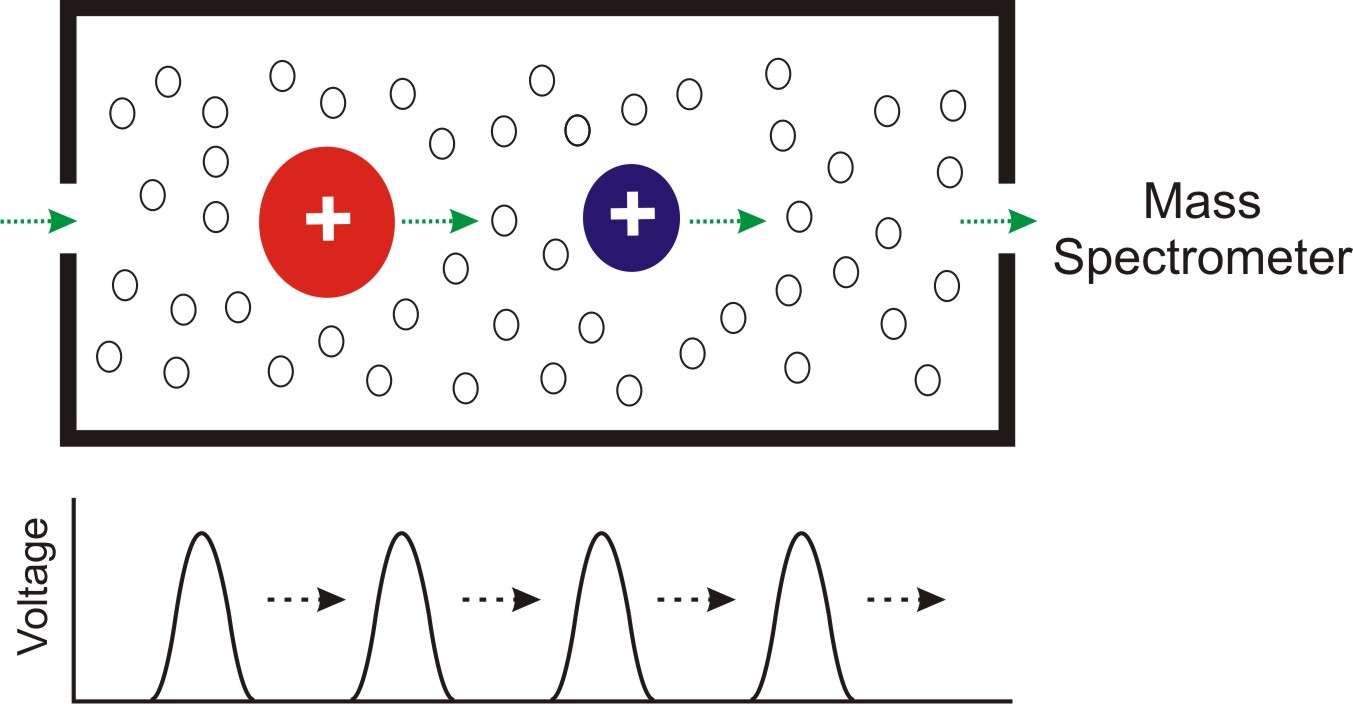
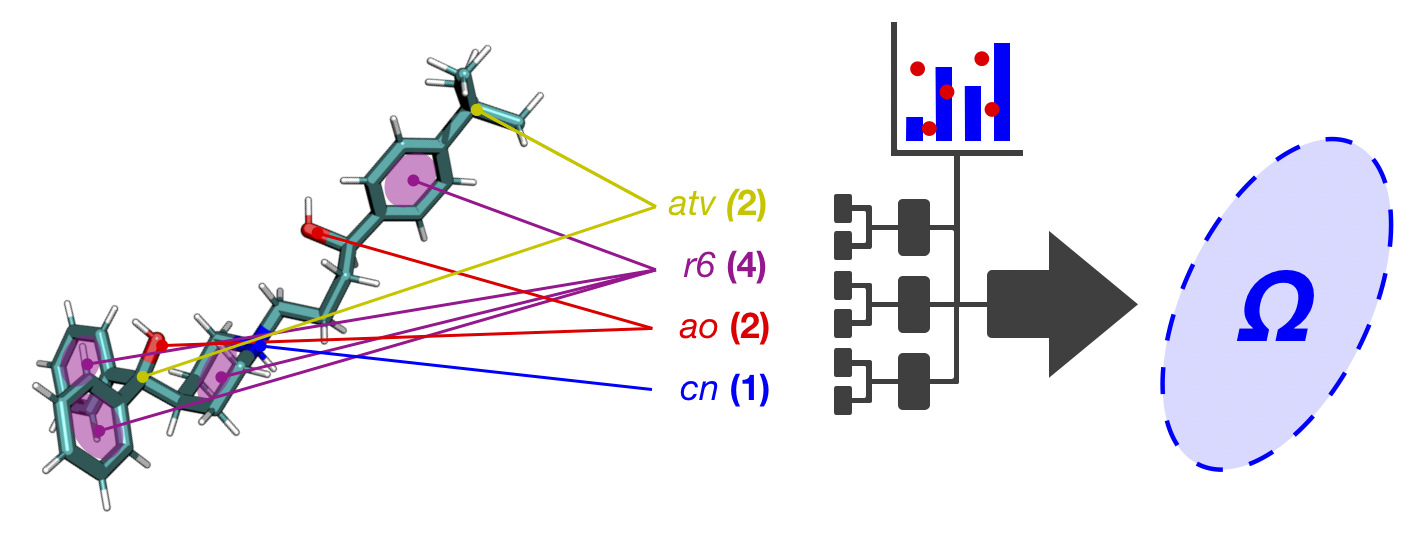
Untargeted metabolomics and lipidomics methodologies are being developed on the platform of the Waters Synapt G2-Si traveling wave ion mobility (TWIM)-QTOF instrument [6]. Ion mobility spectrometry (IMS) separate ions based on the mobility of the ions traveling through a neutral background gas (most commonly helium and nitrogen) under the influence of an electric field. The ion mobility separation is governed by collision frequency between the ions and the neutral gas, i.e., the ion-neutral collision cross section (CCS; Ω). The CCS is determined by the size and shape of the ions in the gas phase and the specific neutral gas. IM separation essentially serves as a rapid gas-phase chromatography, on the scale of milliseconds. When ion mobility is coupled with mass spectrometry (IM-MS), a two-dimensional separation is achieved on the basis of the collision cross section-to-charge (Ω/z) and the mass-to-charge (m/z), respectively [7]. When IM-MS is coupled with UPLC or other chromatographic techniques, a three-dimensional separation is achieved based on retention time (from UPLC), drift time (from IM), and m/z (from MS). The three-dimensional data allows the most definite identification of a specific metabolite. The Figure below illustrates the concept of TWIM-MS, where a migrating voltage pulse push the ions through the neutral gas. Click here for more information.
CCS Database (CCSbase.net)
We have assembled a comprehensive CCS database covering a wide range of chemical space and built a high-performance machine learning-based CCS prediction model for a given SMILES structure. To allow easy access of the database and the prediction model, we have assembled the combined CCS database and the prediction model into a convenient web interface. Check out the database at https://CCSbase.net. Relevant publications: Ross et al. 2020; Ross et al. 2022.
References
1. Porter, F. D., and Herman, G. E. (2011) Malformation syndromes caused by disorders of cholesterol synthesis, J. Lipid Res. 52, 6-34.
2. Xu, L., Davis, T. A., and Porter, N. A. (2009) Rate Constants for Peroxidation of Polyunsaturated Fatty Acids and Sterols in Solution and in Liposomes, J. Am. Chem. Soc. 131, 13037-13044.
3. Xu, L.,* Korade, Z., Rosado, D. A., Mirnics, K., Porter, N. A.* (2013) Metabolism of oxysterols derived from nonenzymatic oxidation of 7-dehydrocholesterol in cells, J. Lipid Res. 54, 1135-1143.
4. Shinkyo, R., Xu, L., Tallman, K. A., Cheng, Q., Porter, N. A., and Guengerich, F. P. (2011) Conversion of 7-dehydrocholesterol to 7-ketocholesterol is catalyzed by human cytochrome P450 7A1 and occurs by direct oxidation without an epoxide intermediate, J. Biol. Chem. 286, 33021-33028.
5. Goyal, S., Xiao, Y., Porter, N. A., Xu, L.,* Guengerich, F. P.* (2014) Oxidation of 7-Dehydrocholesterol and Desmosterol by Human Cytochrome P450 46A1 J. Lipid Res. 55, 1933-1943.
6. Pringle, S. D., et al. (2007) An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-ToF instrument, Int. J. Mass Spectrom. 261, 1-12.
7. Fenn, L. S., and McLean, J. A. (2008) Biomolecular structural separations by ion mobility-mass spectrometry, Anal. Bioanal. Chem. 391, 905-909.