 Cell-cell communication is critical for stomatal patterning Cell-cell communication is critical for stomatal patterning
Despite the fact that epidermal cells in Arabidopsis continue to undergo asymmetric divisions and stomatal differentiation, mature stomata are almost never formed adjacent to each other (Fig. 3). It is important that two stomata are separated at least one cell apart for proper stomatal physiology, as guard cells open and close by osmosis, and necessary water and ions are supplied by adjacent SLGCs. An SLGC almost always divides asymmetrically away from the adjacent, pre-existing stoma (Fig. 3)(Geisler et al. 2000; Geisler et al. 2003). Therefore, cells somehow 'know' the position of existing stomata, and make a fate decision to initiate a stomatal lineage based on this positional information, implying cell-cell communication plays an important role for stomatal patterning.
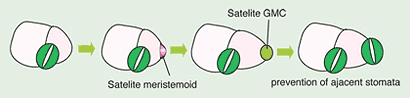
Fig. 3. The 'one-cell spacing rule'. (copy right, Kyoritsu Shuppan, 2006)
Scientists isolated Arabidopsis mutants in which stomatal patterning is disrupted. The loss-of-function mutations in TOO MANY MOUTHS (TMM), STOMATAL DENSITY AND DISTRIBUTION 1 (SDD1), and YODA genes confer plants with elevated numbers of stomata, many of which are clustered (Berger and Altmann 2000; Nadeau and Sack 2002; Bergmann et al. 2004). Their phenotypes suggest that these genes are required to maintain both the number and orientation of asymmetric cell divisions. These gene products define components of cell-cell signal transduction: TMM encodes a transmembrane receptor-like protein with an extracellular leucine-rich repeat (LRR) domain, a protein-protein interaction motif known to act as a cell-surface receptors both in plants and animals (Gomez-Gomez and Boller 2002; Nadeau and Sack 2002; Torii 2004). SDD1 encodes an extracytoplasmic, putative subtilisin-like protease (Berger and Altmann 2000; von Groll et al. 2002). During animal embryogenesis and development, similar proteases are known to modify peptidic ligands (Thacker et al. 1995; Cui et al. 1998). Finally, YODA encodes a homolog of a mitogen-activated kinase kinase kinase (MAPKKK) (Bergmann et al. 2004). MAPKKK acts at the initial point of MAPK cascades, widely-conserved intercellular signaling pathways regulating diverse aspects of cell proliferation and differentiation in plants, fungi, and animals (Marshall 1994; Hirt 1997; Pearson et al. 2001).
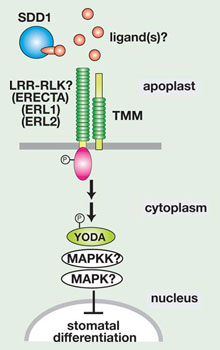
Fig. 4. A hypothetical model of cell-cell signal transduction that enforce 'one-cell spacing rule'. (copy right, Kyoritsu Shuppan, 2006)
A current, prevalent model is that SDD1 modifies yet-unidentified extracellular ligand(s) secreted from developing stomata, which then binds to the TMM receptor in the neighboring cells. This activates the downstream, intracellular signal transduction mediated by YODA- MAPK cascades and in turn suppresses stomatal differentiation (Fig. 4)(Sack 2004). No biochemical experiments have been performed, however, to support this model. Because TMM lacks a cytoplasmic domain, scientists predicted that it must associate with a partner receptor that possesses a cytoplasmic effector domain. Based on previous knowledge, scientists suspected that the partner receptor most likely is an LRR-receptor kinase (Fig. 4). The identity of such a partner, however, remained unclear.
 Synergistic interactions of ERECTA-family receptor-like kinases control stomatal patterning and differentiation Synergistic interactions of ERECTA-family receptor-like kinases control stomatal patterning and differentiation
We discovered that the three ERECTA-family RLKs control stomatal patterning and differentiation. ERECTA, ERL1, and ERL2 together not only promote proliferative cell division, but also suppress asymmetric cell divisions that give rise to stomata. While mutations in each gene do not strikingly affect stomatal patterning, erecta erl1 erl2 plants exhibit excessive asymmetric divisions and formation of high-density stomatal clusters (Fig. 5)(Shpak et al. 2005). Therefore, three ERECTA-family genes act in a synergistic manner to ensure cell-cell communication and fate decisions so that stomata are formed at the right density and in correct positioning. At the same time, each ERECTA-family member possesses a unique function. For instance, ERECTA predominantly suppresses asymmetric cell divisions. In contrast, ERL1 regulates a binary decision of cell fate, a meristemoid vs. an SLGC, after the asymmetric division (Shpak et al. 2005).
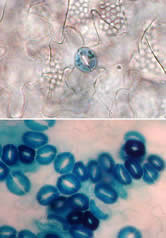
Fig. 5. Loss of three ERECTA-family genes confers high-density stomatal clusters. Top, wild type; bottom, erecta erl1 erl2 triple mutatnt epidermis. (copy right, Junglecity.com, 2005)
Both TMM and three ERECTA-family genes control proper stomatal patterning. TMM is an LRR receptor-like protein without a cytoplasmic effector domain, while the ERECTA-family proteins possess both an LRR and a cytoplasmic kinase domain. Do they form receptor heterodimers that perceive and transmit cell-cell signals to restrict stomatal differentiation? While the exact molecular nature of TMM-ERECTA-family interactions is yet to be revealed, genetic studies have demonstrated the complex interactions among these four receptors. Based on the phenotypes of tmm erecta-family combinatory mutants, it appears that TMM restricts the activity of ERECTA-family RLKs at the initial asymmetric division, satellite asymmetric division, and at the GMC differentiation (Shpak et al. 2005). The interactions depend on the stoichiometry of gene products, suggesting that these gene products act at a close proximity. Perhaps, formation of different combinations of homodimers and heterodimers allows for a delicate balance of cell-cell signals, which leads to correct placing of stomatal complex in a changing environment.
References
Berger, D. and T. Altmann (2000). Genes & Dev. 14, 1119-1131
Bergmann, D. C., W. Lukowitz and C. R. Somerville (2004). Science 304, 1494-1497
Cui, Y., F. Jean, G. Thomas and J. L. Christian (1998). EMBO J 17, 4735-4743
Geisler, M., J. Nadeau and F. D. Sack (2000). Plant Cell 12(11), 2075-2086
Geisler, M. J., D. O. Deppong, J. A. Nadeau and F. D. Sack (2003). Planta 216(4), 571-579
Gomez-Gomez, L. and T. Boller (2002). Trends Plant Sci 7, 251-256
Hirt, H. (1997). Trends in Plant Science 2, 11-15
Marshall, C. J. (1994). Curr Opin Genet Dev 4(1), 82-89
Nadeau, J. A. and F. D. Sack (2002). Science 296(5573), 1697-700.
Pearson, G., et al. (2001). Endocr Rev 22(2), 153-183
Sack, F. D. (2004). Science 304(5676), 1461-1462
Shpak, E. D., J. M. McAbee, L. J. Pillitteri and K. U. Torii (2005). Science 309, 290-293
Thacker, C., K. Peters, M. Srayko and A. M. Rose (1995). Genes Dev 9(8), 956-971
Torii, K. U. (2004). Int Rev Cytol 234, 1-46
von Groll, U., D. Berger and T. Altmann (2002). Plant Cell 14, 1527-1539
copy right, Keiko U. Torii, 2006
|