 Receptor-like kinases in plants Receptor-like kinases in plants
Development of multicellular
organisms relies on coordinated cell proliferation and differentiation.
In animals, growth factor receptor kinases play key roles in cell
differentiation and development, either by stimulating or inhibiting
cell growth. Recent studies revealed that higher plants also possess
genes coding for putative receptor kinases (Receptor-like Kinases,
RLK). For instance, a completely sequenced Arabidopsis genome
contains over 600 genes encoding RLKs (Shiu and Bleecker, 2001),
suggesting that higher plants, like animals, use receptor kinase
signaling commonly and broadly in responding to vast arrays of stimuli
to modulate gene expressions. Although only a handful of RLKs thus far
are shown to have defined biological functions, their roles in
development, self-incompatibility response, and defense against
pathogens illustrate important and versatile function of the RLK
superfamily. However, given that only a few RLKs have been shown to
regulate developmental processes, it is far from being understood how
receptor-kinase signaling control cell proliferation in plants.
A common feature of these
putative receptor kinases (RLKs), is that each has an N-terminal
signal sequence, an extracellular domain that varies in structure,
a single membrane-spanning region, and a cytoplasmic protein kinase
catalytic domain. Unlike animals, where a majority of the receptor
kinases possess tyrosine kinase activity, all of the plant RLKs
thus far are shown to phosphorylate serine-and threonine residues,
except one that displays dual specificity in vitro (Walker, 1994;
Torii and Clark, 2000,; many other great reviews are available!).
Plant RLKs are classified
into subfamilies based on the structural feature of the extracellular
domain, which is thought to act as a ligand-binding site.
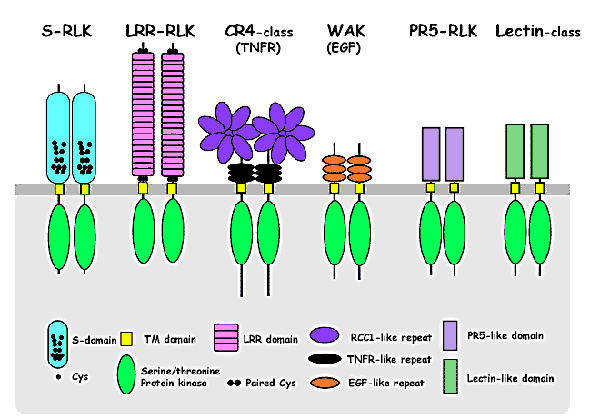
1. S-domain class: S-RLKs possess
an extracellular S-domain homologous to the self-incompatibility-locus
glycoproteins (SLG) of Brassica oleracea (Nasrallah et
al. 1988). The S-domain consists of 12 conserved cysteine residues
(ten of which are absolutely conserved). In addition, the S-domain
possesses the PTDT-box, which has a conserved WQSFDXPTDTFL sequcnce
(X=non conserved amino acid; F=aliphatic amino acid). In Brassica, the S-RLK gene is physically linked to the S locus (Nasrallah et al. 1988). It has been shown
that the S-RLK primarily functions as a receptor for the pollen-derived
ligand, SCR (S-locus cysteine rich protein) during the
self-incompatibility recognition process between pollen and stigma.
The SLG protein is required for a full manifestation of the self-incompatibility
response (Takasaki et al., 2000). However, isolation of
several S-RLK genes from self-compatible plant species
and their expression in vegetative tissues indicate that S-RLKs
may play a developmental role in addition to self-incompatibility.
In addition, one of the S-RLKs of Brassical is implicated
in plant defense response (Pastugalia et al., 1997).
2. LRR class: To date, LRR-RLKs comprise the largest class
of plant RLKs. LRRs (leucine-rich repeats) are tandem repeats
of approximately 24 amino acids with conserved leucines. LRRs
have been found in a variety of proteins with diverse functions,
from yeast, flies, human, and plants, and are implicated in protein-protein
interactions. Several LRR-RLKs have been shown to play critical
roles in development. Those include ERECTA which regulates organ
shape, CLAVATA1 which controls cell differentiation at the shoot
meristem, HAESA, which regulates floral abscission process, and
BRI1, which is involved in brassinosteroid perception (Torii et al., 1996; Clark et al., 1997; Li and Chory 1997;
Jinn et al., 2000). On the other hand, the rice gene Xa21 confers race-specific resistance to Xanthomonas oryzae pv oryzae (Song et al., 1995). Therefore, LRR-RLKs also play a role in disease
resistance. Interestingly, the tomato Cf disease resistance
gene products, which confer a race-specific resistance to Cladosprium
fulvum, contain extracellular LRR domains but lack the cytoplasmic
protein kinase domain (Jones and Jones 1997).
3. TNFR class: The maize CRINKLY4 (CR4)
gene product possess TNFR (tumor-necrosis factor receptor)-like
repeats, that has a conserved arrangement of six cyeteines, and
seven repeats of ~39 amino acids that display a weak similarity
to the RCC GTPase (Becraft et al., 1996, McCarthy and Chory,
2001). CR4 is required for a normal cell differentiation
of the epidermis (Becraft et al., 1996). The Aragbidopsis genome contains several genes related to CR4 (McCarthy and Chory, 2001).
4. EGF class: The cell wall associated receptor kinases
(WAKs) represent the EGF (epidermal growth factor) class. The
EGF-like repeat motif is characterized by a conserved arrangement
of six cysteines. The EGF-like repeats are found in variety of
animal extracytoplasmic receptor domains and are known to play
a role in protein-protein interactions. In Arabidopsis,
four WAKs (WAK1 to WAK4) have been identified, and all of them
have extracellular EGF-like repeats (He et al., 1996; Ellard-Ivey et al., 1997). Reverse-genetic experiments suggest that WAKs
may be involved in pathogenic responses (He et al., 1998).
5. PR class: The Arabidopsis PR5K (PR5-like
receptor kinase) is the known example of this class. The extracellular
domain of PR5K exhibits sequence similarity to PR5 (pathogenesis
related protein 5), whose expression is induced upon pathogen
attack (Wang et al., 1996). The structural similarity between
the PR5K receptor domain and PR5 suggests a role for PR5K in pathogenesis
response.
6. Lectin class: The Arabidopsis LecRK1 gene
product possesses an extracellular domain homologous to carbohydrate-binding
proteins of the legume family (Harvé et al., 1996).
Although biological function of LecRK1 is yet known, its structural
feature suggests that LecRK1 may be involved in a perception of
oligosaccharide-mediated signal transduction. The Arabidopsis genome contains >30 genes belonging to Lectin-RLKs several
genes coding for Lectin-RLKs (McCarthy and Chory, 2001).
7. Others: The RLKs, which possess extracellular domains sharing no homology to known motifs, are classified as "Others".
References
Becraft, P. W., et al. (1996). Science 273: 1406-1409.
Clark, S. E., et al. (1997). Cell 89: 575-585.
Ellard-Ivey, M., T., et al. (1997). Plant Cell 9: 2105-2117.
Harvé, C., P., et al. (1996). Journal of Molecular Biology 258: 778-788.
He, Z.-H., et al. (1996). Journal of Biological Chemistry 271: 19789-19793.
Jinn, T., et al. (2000). Genes & Dev. 11: 108-117.
Jones, D. A. and J. D. G. Jones (1997). Advances in Botanical Research incorp Advances in Plant Pathology 24: 90-167.
Li, J. and J. Chory (1997). Cell 90: 929-938.
McCarty, D. R. and J. Chory (2000). Cell 103(2): 201-9.
Nasrallah, J. B. (2000). Curr Opin Plant Biol 3(5): 368-73.
Nasrallah, J. B., et al. (1988). Proc Natl Acad Sci USA 85: 5551-5555.
Pastuglia, M., D., et al. (1997). Plant Cell 9: 49-60.
Shiu, S.H. and Bleecker, A.B. (2001) Proc Natl Acad Sci USA 98: 10763-10768.
Song, W.-Y., et al. (1995). Science 270: 1804-1806.
Takasaki, T., et al. (2000) Nature 403: 913-916.
Torii, K. U. and S. E. Clark (2000). Advances in Botanical Research incorp Advances in Plant Pathology. 32: 226-268.
Torii, K. U., et al. (1996). Plant Cell 8: 735-746.
Walker, J. C. (1994). Plant Molecular Biology 26: 1599-1609.
Wang, X., et al. (1996). Proc Natl Acad Sci USA 93: 2598-2602.
© Keiko U. Torii, 2004
|