
Bioengineering Department, Box 355061, University of Washington, Seattle, WA 98195, USA

 |
Bioengineering Department, Box 355061, University of Washington, Seattle, WA 98195, USA |
 |
Return to Yager's Home Page
|
|
Delivery of therapeutic agents to the intended site of action at a controlled rate is often difficult. We have been exploring the potential of lipid complex high axial ratio microstructures (CHARMs) as components of continuous delivery systems for such agents. CHARMs include lipid tubules, helices (like those shown in a darkfield optical micrograph at left), and cochleate cylinders. In collaboration with Michael Gelb of Chemistry, we synthesized dozens of lipopeptides and other surfactants that self-assembled into CHARMs and could associate with therapeutics. We found that covalent attachment of therapeutic molecules to the headgroups of lipopeptides that formed CHARMs could greatly change the bioavailability of that therapeutic. We engaged in 3 in vivo testing collaborations: 1) with Mary L. (Nora) Disis in Oncology to explore the potential for these microstructures as vaccines against cancer and infectious disease, 2) with John Amory of the VA Hospital to explore CHARM-based delivery of testosterone, and 3) with Peter Tarcha of Abbott Research Laboratories. Project completed. |
More Information on This Project
|
|
Since 1994 we have been designing microfluidic devices and systems for use in monitoring the physical and chemical nature of complex fluids such as blood. The work was initially supported by the Washington Technology Center, DARPA DSO and Senmed Medical Ventures, Micronics, Inc., of Redmond, WA, a company founded on the basis of intellectual property developed in this project. We are currently engaged in expanding the our understanding of the capabilities of the T-sensor, a version of which is shown at left. We are also making a transition from Si microfabrication to materials amenable to use in inexpensive disposable devices. While this project is officially completed, similar work is ongoing in the laboratory (see Current Work). |
More Information on This Project
A central problem in the development of Microfluidic Molecular Systems is that while many excellent methods exist for detecting and quantifying chemical and biological warfare (CBW) agents (some of which have already been miniaturized to the MEMS size range), the macroscopic sample preparation methods required to continuously extract the analytes from the extraneous matter in "real world" samples prior to chemical measurements have not been miniaturized.
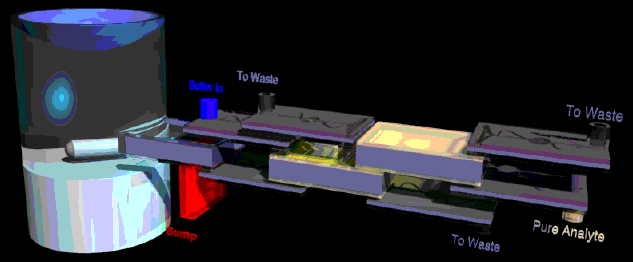
As shown in the (highly schematic) working drawing above, we worked to design a system that could plug directly into an air sampler, incorporating pumps (thin devices) and flow control modules interconnecting three sequential microfluidic components: a sedimentation device at left (connected to the red sump), an isoelectric focusing element (gold electrodes showing at the center), and an electrophoretic concentrator (at right, also with gold electrodes). Our project was to develop a microfluidic system that would allow sorting of analytes prior to chemical identification.
The multidisciplinary project was funded by the DARPA MTO MicroFlumes program from May 1997 to July 2000. It had the following aims:
1. Development of a chemical separation system that takes advantage of low-Reynolds number conditions present in microfabricated fluid channels. (Yager Group)2. Development of a sample pretreatment system that allows extraction of the relevant analytes from fluids containing interfering non-analyte particles and concentration of analytes initially collected at low concentrations in large volumes of fluid. (Yager Group)
3. Development of on-chip pumping systems that are tolerant of fluids containing particles of widely differing sizes. (Forster Group)
The project was a collaboration between professors Yager, Fred K. Forster (ME), and Martin A. Afromowitz (EE) at UW. Work in the Yager lab focused on the development of sedimentation, electrophoresis and isoelectric focusing as methods for sample fractionation. Several novel microfluidic devices and methods have resulted.
More Information on the Yager Lab component of This Project
![[model of silk beta sheet crystallite]](silkcpk.gif) |
Natural silks match many of the properties of high-performance synthetic polymers, yet are processed under significantly milder conditions. High strength, stiffness and impact resistance are achieved in a polymer that is precipitated from aqueous solution at room temperature; also, this material is biodegradable, producing nontoxic breakdown products. A variety of natural silk secretions form liquid crystalline phases en route to solidifying. This project, now complete, was aimed at developing a link between the folding of proteins and liquid crystalline polymer technology through an examination of the molecular and microstructural changes that accompany the spinning of silk fiber. Microstructural and molecular changes were observed in both natural and artificial silk spun through orifices. Key personnel included Kimberly Carlson (nee Trabbic). |
|
|
This project was supported as a subcontract to UW from MesoSystems Technologies. It was ultimately supported by the U.S. Army. It was the most recent in a series of projects we have had supported by DARPA and MesoSystems aimed at improving detection of chemical and biological warfare agents through application of microfluidics. There were two overall tasks to be performed in this project. The first was to determine whether a new design for application of high voltage to microfluidic channels could be used effectively for zone electrophoresis and isoelectric focusing. The second was to apply the best possible method to developing an electrokinetic method for isolation of DNA from bacterial cells. That aspect of the work was to be developed in two stages. In the first phase, the input sample was to consist only of dilute DNA, including DNA from a BW simulant such as Bg. The aim as to be to demonstrate and quantify concentration of this material using either zone electrophoresis or isoelectric focusing. The goal was to be concentration of DNA by a factor of 10. In the second effort, the initial sample was to be vegetative bacteria. Upstream of the isoelectric focusing step, bacteria were to be lysed using detergents. |
More information on MesoSystems
|
|
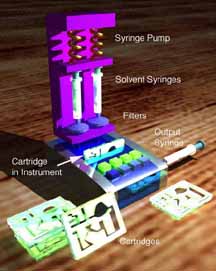
This project was supported the Hewlett Packard Corporation through their thermal inkjet printer group in Corvallis, OR. Proposed was a preliminary collaborative project to determine if HP’s thermal inkjet (TIJ) printer technology could be adapted to the printing of protein arrays useful to the Yager laboratory, and, by extension, useful to the research and medical diagnostic communities. We evaluated the ability of said TIJ printers 1) to eject a small set of representative proteins in functional form, 2) to eject protein solutions without excessive loss of protein to the walls of the ink jet cartridges, and 3) to form small functionalized regions on representative surfaces using thiols and proteins. At left is a false-colored image of the result of a first attempt at a surface plasmon resonance imaging immunoassay using a TIJ-printed surface. The technology was used in a preliminary demonstration of a novel immunoassay using the imaging technique. |
|
|
One of the most powerful and versatile biomedical diagnostic
tools, the immunoassay, is used to monitor the levels of drugs and hormones
in body fluids, to diagnose infectious and autoimmune diseases, and
to both diagnose and monitor treatment of cancer. The performance of
immunoassays is today largely restricted to centralized laboratories
because of the need for long assay times, complex and expensive equipment,
and highly trained technicians. If a wider range of the 700 million
immunoassays performed annually in the US alone could be run more inexpensively,
more frequently, and at the point of care, the health of millions of
patients could be improved. Recent developments in microfluidics suggest
that instruments could soon be developed that would allow immunoassays
to be performed as easily as is blood glucose testing today. |
|
|
The collection of saliva is far preferable to collection of blood from the point of view of the person being sampled. However, in recent years microfluidic technologies for measuring analytes in blood have advanced rapidly, while the use of saliva as an analyte has lagged, both in terms of the number of analytes measured and the environments in which such measurements are made. In part, this is because saliva is more variable than plasma, has analytes in lower concentrations, and contains viscous and adhesive mucins. If it were practical to use saliva for many analytes commonly measured in blood, and to make those measurements on several analytes at once, inexpensively, and in a way not requiring technical training, enormous improvements in the quality, frequency and scope of biomedical testing for research, therapy, and health maintenance would be possible, particularly for ambulatory outpatients. This project developed integrated microfluidic systems for rapidly, inexpensively, and simultaneously measuring multiple analytes in saliva, and in a simple disposable polymeric laminate format. A microfluidic device to allow rapid extraction of analytes from the mucins in saliva was developed. Two new but demonstrated immunoassay technologies were coupled to a microfluidic system that allowed dry storage of all reagents at ambient conditions and measured multiple analytes in parallel. These assays can measure low levels of hormones, drugs, metabolites, and even proteins that indicate the presence of disease, as well as compounds specific to the oral cavity such as pathogens and markers for oral cancer. The immunoassays were initially be validated on hormones for which commercial immunoassays were available. Ongoing work on development of parallel diffusion immunoassays were extended to saliva testing through coupling with the mucin-extraction system. To measure analytes present at concentrations below the limit of detection of the diffusion immunoassay, chemically amplified surface plasmon resonance (SPR) imaging was used. The design of a novel SPR microscope developed in this project is shown at left. Ultimately, three microfluidic assays were developed with promise to detect a range of analytes in this or related optical readout formats. |
|
This project was supported by the Singapore government's A*STAR, through the Singapore University of Washington Alliance (SUWA). My group collaborated with Prof. Tjin at Nanyang Technological University, whose work focused on development of compatible instrumentation. |
The goal of this project was to develop an inexpensive portable assay system for monitoring multiple biochemicals in blood simultaneously and in a few minute at the point of care. The system was to be based on a simple microfluidic disposable component containing all chemicals required for the bioassays that can be stored indefinitely at ambient conditions. Work included development of: appropriate assay chemistry that can be stored dry, disposable laminates that can support several assays types and the storage requirements, a process for reconstituting the assay chemistries after long-term storage (shown schematically on the left), and the low-cost portable instrumentation to read and interpret the assay results. UW focused on the dry reagent technology, microfluidic laminates and fluorescence imaging of enzymatic assays in solution and surface-bound fluorescence immunoassays. |
|
The Longmuir laboratory at UC Irvine developed a non-viral gene delivery system strategy based upon zwitterionic lipid, peptides, and polymer components. Each component can be individually optimized to overcome the sequential barriers to gene delivery, which are 1) assembly, 2) extracellular stabilization, 3) endosomal/lysosomal escape, and 4) nuclear entry in the absence of cell division. The delivery system is assembled in 25% ethanol/75% water mixtures, avoiding the use of toxic organic solvents and detergents. Under subcontract to UC Irvine as part of a 5-year NIH grant, the Yager group worked to establish microfluidic technology that could lead to fully automated assembly of non-viral gene delivery complexes (as shown schematically at left). The steps explored were:
This work has led to novel approaches to nanoparticle formation, and control of the process by modifying the solutions used in rapid mixing. |
|
|
Diarrhea is one of the major causes of death in the developing world, particularly among children. Even in the developed world, the standard method for determining the cause of diarrhea is culturing for bacteria, which can take up to 3 days. A rapid test for the pathogen responsible for diarrhea would, for the first time, allow tailoring treatment for particular patients. The Seattle-based nonprofit organization PATH obtained a grant from NIH/NIAID to develop a system for measuring the levels of pathogens in human stool samples for the rapid diagnosis of the causes of diarrhea. Subcontractors under the leadership of PATH's Bernhard Weigl were Micronics of Redmond, WA, Dr. Philip Tarr of Washington University in St. Louis, and the Yager group. The final instrument developed by Micronics utilized rapid on-card PCR for identifying the pathogenic strain. The Yager group was responsible for developing a system for drying, preserving, and rehydrating the reagents required. Shown at left is a preliminary phase diagram for the fluorescence emission of a dye used to monitor the degree of dryness in small samples of the stabilizing matrix. |
|
|
The Yager group was part of a collaboration that included the Jet Propulsion Laboratory in California that was led by Prof. David Stahl of the UW Department of Civil Engineering and funded by NASA. The aim of the project was to develop a nucleic acid probe microarray-based sensor for the detection and evaluation of microorganisms that contaminate the International Space Station drinking water system. Using this sensor we would not only monitor the microbial contamination, but would identify and quantify the problematic microbial species (e.g., opportunistic pathogens) in the ISS drinking water that pose a health risk to astronauts. The approach was to measure the RNA content of lysed microbes using fluorescence monitoring of a gel-based microarray. The Yager group contribution to the project was converting the current sample preconditioning and processing steps to a microfluidic format. The requirement for low instrument weight and operation in the absence of gravity added some interesting challenges. Image courtesy of NASA. |
|
A Seattle-based consortium led by the Yager group was awarded $15.4 million by the Bill & Melinda Gates Foundation's Grand Challenges in Global Health initiative in 2005. The aim of the project was to develop a portable device that would bring the technological power of a modern medical diagnostics laboratory to the developing world. The consortium coinvestigators were Patrick Stayton of the University of Washington Department of Bioengineering, Gonzalo Domingo of PATH, Fred Battrell of Micronics, Inc., WA, and Walt Mahoney of Epoch Biosciences Inc (as of 2016 part of the ELITech Group). Developing countries have limited resources and lack facilities to test patients whose symptoms indicate they may have a life-threatening infectious disease. The consortium's efforts were directed toward filling the need for an affordable, portable system that does point-of-care tests for multiple analytes from a single sample and provides results in a matter of minutes. The project developed and tested a prototype of an instrument--the DxBox (at left)--that healthcare workers could bring to remote areas of the developing world to quickly and easily make diagnoses. The chemical processes--immunoassays and PCR-based nucleic acid amplification--were carried out on disposable plastic lab cards that contain all the reagents, can be stored at ambient conditions for a year, and use a few drops of blood as the sample. The initial clinical target was a panel of tests that differentiate between six pathogens that are likely to cause fever in the developing world. Micronics, Inc. is responsible for commercializing the DxBox after the end of the project in November of 2010. This has not occurred as of July 2017. Micronics is now a wholly-owned subsidiary of Sony Corporation. As of the sale of Micronics to Sony, Yager has no financial interest in the company. |
|
This 2-year project was funded by NIH/NIBIB through the ARRA Challenge Grant mechanism. The aim was to demonstrate the capabilities of a two-dimensional paper network (2DPN) to perform complex sequences of chemical processes resulting in enhanced detection of biomolecules. The 2DPN format was 1) instrument-free, 2) operatable by an untrained user, 3) allowed fully quantitative multiplexed analyte measurement using a simple camera, and 4) inexpensive enough to be disposable. In essence, it was to be the DxBox where the box will be replaced by a cell phone. A multiplexed diagnostics platform that has higher sensitivity and greater functionality than current lateral flow tests, but at a cost-per-test and with an ease-of-use comparable to those tests, has the potential for very positive impact on human heath. This type of device could be of use to diagnose a number of health conditions in many low-resource settings in the U.S. and the developing world. In the developed world, the biggest impact would be for use in physicians’ office laboratories, nursing homes, hospitals, and in the home. Potential high impact applications are those in which there is a need for significantly improved detection sensitivity to enable earlier diagnosis (and treatment) than is possible with current lateral flow tests. These include multiplexed testing for the full range of infectious agents, as well as biomarkers for acute and chronic disease. The project was funded in 2009 and terminated on schedule in August of 2011. |
|
This was a 9-month DARPA DSO "seedling project", the aim of which is to demonstrate that the 2DPN platform can be used for rapid point-of-care detection of emerging diseases. The 2DPN is a sensitive instrument-free multiplexed diagnostic platform using disposable devices that are comparable in cost and ease of use to conventional lateral flow tests. Based on previous experience, we chose detection of dengue virus as an initial target. Dengue is an emerging infectious disease that infects over 50 million people each year in 100 countries (with recent cases being reported in Hawaii, Texas, and Florida), and is classified by NIAID as a Group III pathogen with bioterrorism potential. The demonstration of the platform was to be a multi-analyte immunoassay for the four serotypes of dengue virus, as well as for previous dengue infection (IgG). The platform was to be applicable to other targets including other emerging infectious diseases that could affect deployed military personnel. The anticipated greater sensitivity and multiplexing of the proposed 2DPN platform are improvements over conventional lateral flow tests for the effective early diagnosis of dengue (e.g. identification of previous infection increases the risk of developing dengue hemorrhagic fever). Dates of performance: 10/10 – 6/11 |
A Seattle-based consortium led by the Yager group was awarded $5.7 million in 2011 by the NIAID (an NIH institute). The aim of the 5-year project was to develop a portable device that would increase the sensitivity of protein immunoassays with a combination of using 2DPNs and novel protein binders, with the specific goal of producing a more sensitive assay for influenza. |
The dual goals of the project were: 1) to increase the sensitivity of the conventional influenza A/B nucleoprotein immunoassay t through chemical amplification of the detection binding event using the HRP-DAB system as implemented by a 2DPN, and 2) to develop novel high-affinity stable protein binders (as an alternative for antibodies) for the HA protein that allow implementation of an HRP-DAB amplified sandwich assay for HA or intact virus on a 2DPN. A test of a nucleoprotein assay prototype device (see image) for assay of nasal swabs from patients at an ER was carried out at Seattle Children's hospital during the 2015-2016 flu season. The consortium coinvestigators included David Baker of UW's Department of Biochemistry, David Moore of the GE Global Research Center, Elain Fu of UW (now of Oregon State University), Gonzalo Domingo of PATH, and Janet Englund of Seattle Children's. |
A Seattle-based consortium led by the Yager group was awarded $19.7 million in 2011 by the DARPA Defense Sciences Office (DSO, later transferred to BTO). The aim of the 5-year project was to develop an instrument-free fully-disposable paper-based diagnostic platform capable of detecting multiple pathogens by their DNA or RNA from a patient sample. |
Performance goals for the MAD NAAT device included storage for 1 year at ambient conditions (up to 45°C), incorporation of all reagents on the stored device, and sample-to-result operation within 1 hour. The initial test was to identify MRSA from a nasal swab sample (using DNA), then RSV (using RNA). In the last year of the project the target pathogens were expanded to include Chlamydia trachomatis and Neisseria gonorrhoeae (DNA) in urine and the Zika virus in blood. The device was to go from sample-to-result anywhere, be simple enough for untrained users, and low enough cost to enable wider use of sophisticated medical tests. Potential high-impact applications are those in which there is a need for significantly improved detection sensitivity to enable earlier diagnosis (and treatment) than is possible with current protein-based tests. These include multiplexed testing for the full range of infectious agents in human and environmental samples. The consortium coinvestigators included Barry Lutz of UW Bioengineering, Walt Mahoney of Epoch Biosciences (later the ELITech Group), David Moore of the GE Global Research Center, Bernhard Weigl of PATH, Ferric Fang of UW Laboratory Medicine, and Janet Englund of Seattle Children's. |
A color image of a grid of spots of green-colored fluorescein and red-colored Texas Red on a glass fiber substrate at concentrations relevant to point-of-care diagnostic tests. Fluroescence image captured by a mobile phone camera with a dual-wavelength excitation filter on the camera flash and a dual-wavelength emission filter on the camera simultaneously images two fluorophores such as |
This was a 2-year NSF EAGER project (and NCE) aimed at utilizing the full potential of current and future smart phones for medical applications, particularly those involving optical detection of biochemical assays. The principal aim was to determine what optical capabilities should be incorporated into the next generation of smart phones to be useful in medical diagnostics tests. Initially this project was aimed at improving the sensitivity of lateral flow tests by utilizing the photoacoustic effect. The conventional lateral flow test (as in a commercial home pregnancy test) uses a dark optical label like a Au nanoparticle to create a couple of lines on a white (scattering) material like a porous nitrocellulose membrane. If there are fewer than about 10E8 nanoparticles in a detection line, the human eye cannot detect them. The idea was to use a time-modulated laser to heat the Au nanoparticles enough to generate a brief "click" that could be picked up by a microphone on or near the lateral flow strip. Signals proved to be too small. The rest of the project aimed at using the camera of a cell phone to collect signals from low-concentration fluorophores to ehnahce lateral flow test sensitivity, which did work. Early work was performed in collaboration with Prof. Matthew O'Donnell, and the latter work in the Yager lab was performed by Peter Kauffman and Ph.D. candidate Kamal Shah. |
|
David Baker's group led an R21 project aimed at developing enhanced-specificity serology assays that could differentiate the presence of an ongoing or pre-existing immune reponse to closely-related flaviviruses, particularly Zika, dengue and yellow fever viruses. There were two primary goals of this project. 1) The Baker group was to develop the molecular tools (small novel viral epitope mimetic proteins) capable of selectively capturing and detecting just those antibodies that are specific to the target viruses. 2) The Yager group was to integrate the novel binders into a simple low-cost rapid 2DPN-based point-of-care serology assay based on binding the target antibodies from a plasma sample. The project ultimately failed to produce appropriate binders, although there was good progress on the device design and testing. |
In collaboration with the David Baker group, we were awarded $4.3 million in 2016 by the Defense Threat Reduction Agency (DTRA). The aim of the 2-year contract was to develop an instrument-free fully-disposable paper-based diagnostic platform capable of extremely sensitive detection of Ebola viral glycoproteins from blood. {TEM of EBOV, Oct. 13, 1976, by, Frederick A. Murphy of the CDC} |
There were three primary goals of this project. 1) Develop a hyper-sensitive protein assay and embody it in a rapid POC disposable format using 2DPN technology. 2) Develop the molecular tools (novel binding proteins) for capturing and detecting glycoproteins from Ebolavirus in a sandwich assay on paper. 3) Integrate both technologies into an disposable that detects EBOV. Some progress was made in developing the binders to Ebolaviral proteins, and a way was found to attach them to paper for a lateral flow assay. It was also demonstrated that there was great promise in enhancing the sensitivity of protein assays in lateral flow by using nucleic acid templates on the detection binders, followed by a nucleic acid amplification step. The components were never fully integrated into a viable device. |
|
Return to Yager's Home Page Return to Research Page |