Biological endpoints Evaluating the radiation effects on a cell system is achieved by measuring one or more of the final outcomes that occur to the system, which are called biological endpoints. The surviving fraction, the number of unrejoined DSB and neoplastic transformation are three biological endpoints of special interst interest within the VC system of models. Surviving Fraction Surviving fraction (S or SF) is the fraction of the cells that are able to produce viable progeny at a given time point in a cell culture, expressed as a ratio to the number of reproductively viable cells divided by the number of irradiated cells. What is FAR? DSB rejoining rate data are often reported in the earlier literature in terms of FAR (% activity out of the plug). The random breakage model (Contopoulou et al. 1987, Cook and Mortimer 1991) provides a suitable formalism to convert model predictions of the number of unrejoined DSBs into an estimate of FAR. The equation relating unrepaired DSBs to the FAR is (Belli et al. 2000) 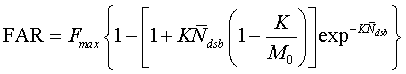 Here, Fmax is the maximum fraction of the cellular DNA that can enter the gel, M0 is the average chromosome size (in base pair), Ndsb is the number of radiation-induced DSBs, and K is the gel exclusion size (in base pair). DNA fragments larger than K do not move out of the well. In general, the fraction of the DNA migrating out of the agarose plug (well) tends to increase as the gel exclusion size increases and decreases as the average chromosome size increases. Neoplastic cell transformation Normal cells exposed to carcinogenic agents such as radiation, acquire altered characteristics in morphology (structure of the cell), metabolism and growth in vitro, and the they may exhibit tumor formation in vivo. These phenotypic changes are called transformations. There is another form of cell transformation called "spontaneous transformation", a phenomenon observed in vitro when maintained for a considerable period of time. Transformation is hardly a one-step process, but consists of multiple stages that involves a number of successive cell generations allowing each progeny to increasingly deviate until the final irreversible stage of transformation is reached. Full transformation to the neoplastic state is denoted by the character that they can produce tumors in vivo (when transplanted to an appropriate host). Here, Fmax is the maximum fraction of the cellular DNA that can enter the gel, M0 is the average chromosome size (in base pair), Ndsb is the number of radiation-induced DSBs, and K is the gel exclusion size (in base pair). DNA fragments larger than K do not move out of the well. In general, the fraction of the DNA migrating out of the agarose plug (well) tends to increase as the gel exclusion size increases and decreases as the average chromosome size increases. Neoplastic cell transformation Normal cells exposed to carcinogenic agents such as radiation, acquire altered characteristics in morphology (structure of the cell), metabolism and growth in vitro, and the they may exhibit tumor formation in vivo. These phenotypic changes are called transformations. There is another form of cell transformation called "spontaneous transformation", a phenomenon observed in vitro when maintained for a considerable period of time. Transformation is hardly a one-step process, but consists of multiple stages that involves a number of successive cell generations allowing each progeny to increasingly deviate until the final irreversible stage of transformation is reached. Full transformation to the neoplastic state is denoted by the character that they can produce tumors in vivo (when transplanted to an appropriate host). References - M. Belli, R. Cherubini, M. Dalla Vecchia, V. Dini, G. Moschini, C. Signoretti, G. Simone, M.A. Tabocchini, P. Tiveron. DNA DSB induction and rejoining in V79 cells irradiated with light ions: a constant field gel electrophoresis study. Int. J. Radiat. Biol. 76(8):1095-104 (2000).
- C.R. Contopoulou, V.E. Cook and R.K. Mortimer, Analysis of DNA double strand breakage and repair using orthogonal field alternation gel electrophoresis. Yeast 3(2), 71-6 (1987).
- V.E. Cook and R.K. Mortimer, A quantitative model of DNA fragments generated by ionizing radiation, and possible experimental applications. Radiat. Res. 125(1):102-6 (1991).
MCAT: NPAR = {number of parameters to adjust}
AMPi: CID={string} PNAM={string} PLB={value} PUB={value} PIG={value} CID: Parameter categoryPNAM: Parameter namePLB: Minimum allowed value for the parameter (parameter lower bound)PUB: Maximum allowed value for the parameter (parameter upper bound)PIG: Initial guess at the optimal parameter value (parameter initial guess)
Note for tutorials 17 and 18 : Measured (experimental) data is used in order to estimate biological parameters for the models LPL, RMR or TLK using VC. The measured biological data such as DSB, FAR (fraction of activity released) or surviving fractions are preprocessed using the utility code XIO and then VC is run to use this data to best fit the model and estimate the relevant parameters through non-linear regression techniques. In this tutorial, the use of utility code XIO, a problem of estimating the several model parameters using measured FAR and using a set of estimated parameters to fit a different set of data for the same biological/analytical system are demonstrated. Data preprocessing using XIO (Tutorial #17) - Example 17.1 Preprocessing DSB damage (FAR) data.
- Example 17.2 Simultaneously preprocessing FAR and survival data.
Estimating biological parameters using measured (experimental) DSB data (Tutorial #18)- Example 18.1 Using measured FAR data to estimate model parameters.
Note for tutorials 19 and 20: Estimating biological parameters using survival data is performed much the same way as using DSB or FAR data (tutorial 17 and 18). The only difference appears in the input files for the XIO application. RMR model is used in Example 19.1 to demonstrate the estimation of three parameters using E. coli data. Data preprocessing using XIO (Tutorial #19)- Example 19.1 Preprocessing E. coli survival data.
Estimating biological parameters using measured cell survival data (Tutorial #20)- Example 20.1 Using measured survival data to estimate RMR model parameters.
|

