Genomic Instability Genomic instability refers to abnormally high rates (possibly accelerating rates) of genetic change occurring serially and spontaneously in cell-populations, as they descend from the same ancestral cell. By contrast, normal cells maintain genomic stability by operation of elaborate systems which ensure accurate duplication and distribution of DNA to progeny-cells, while preventing duplication of genetically abnormal cells (Cheng 1993). These systems ("metabolic pathways") involve an estimated 100 genes (Cheng 1993). The yield of non-lethal genetic alterations (point mutations and chromosome aberrations) produced in a cell by radiation can be plausibly linked to the activation or inactivation of critical genes, the induction of genetic instability, and neoplastic transformation (Coleman and Tsongalis 1995, Hanawalt 1998, Schmutte and Fishel 1999). Through the DNA damage formation and repair models, the VC software can estimate the fraction of the initial DNA damage that is converted to lethal and non-lethal mutations. VC treats the probability that radiation damage causes genome instability to be a function of dose and dose rate.
Neoplastic Transformation Neoplastic transformation is the conversion of normal cells into tumor cells. Frequently this is the result of a genetic change (mutagenesis). After irradiation, a stochastic fraction of the surviving cells is promoted to a state of enhanced genome instability (i.e., stage 1 transformed cells). A stochastic fraction of the remaining viable cells are left in an untransformed state with, perhaps, minor genome instability (see the figure below). As illustrated in the figure, the untransformed and stage 1 transformed cells can transition to a neoplastic state (at different transformation rates). 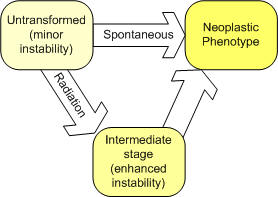
A generalized and concise graphical representation of cellsleading to neoplastic transformation as used in VC. The current version of the VC software uses a multi-target, multi-hit model to link the expected number of non-lethal genetic alterations in a cell to the onset of genetic instability (see the theory section). The models do not explicitly account for apoptosis or bystander effects. References Coleman WB and Tsongalis GJ, Multiple mechanisms account for genomic instability and molecular mutation in neoplastic transformation. Clin. Chem. 41(5), 644-657 (1995). Hanawalt PC, Genomic instability: environmental invasion and the enemies within. Mutat. Res. 400(1-2), 117-125 (1998). Schmutte C and Fishel R, Genomic instability: first step to carcinogenesis. Anticancer Res. 19(6A), 4665-4696 (1999). Cheng KC, Genomic Instability and Tumor Progression: Mechanistic Considerations. Advances in Cancer Research 60: 121-156 (1993).
|
|

