mmf.math.linalg.cholesky.gmw81¶
| gmchol(A[, test]) | Return (L, e): the Gill-Murray generalized Cholesky |
| colmod(A) | Modified Cholesky factorization |
| mchol2(A) | Based on the MATLAB code |
These are a collection of codes based on the [GMW81] algorithm.
Comments¶
- colmod() :
- Seems to work well and is quite efficient as it has been vectorized, but not quite as good as gmw(). Use this for now.
- gmw() :
- Seems to work well, but returns an explicit pivoting matrix and is very slow as it uses loops.
- gmchol() :
- Fast (uses weave) but very bad for large matrices because it uses no pivoting.
- mmf.math.linalg.cholesky.gmw81.gmchol(A, test=False)[source]¶
Return (L, e): the Gill-Murray generalized Cholesky decomposition of M = A + diag(e) = dot(L, L.T) where
- M is safely symmetric positive definite (SPD) and well conditioned.
- e is small (zero if A is already SPD and not much larger than the most negative eigenvalue of A)
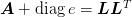
Parameters : A : array
Must be a non-singular and symmetric matrix
test : bool
If True, use the directly translated iterative code for testing.
Returns : L : 2d array
Lower triangular Cholesky factor.
e : 1d array
Diagonals of correction matrix E.
Notes
- Translated from the following Gauss code of J. Gill::
****************************************************************/ * This is the Gill/Murray cholesky routine. Reference: / * */ * Gill, Jeff and Gary King. ``What to do When Your Hessian */ * is Not Invertible: Alternatives to Model Respecification */ * in Nonlinear Estimation,’’ Sociological Methods and */ * Research, Vol. 32, No. 1 (2004): Pp. 54–87. */ * */ * Abstract */ * */ * What should a researcher do when statistical analysis */ * software terminates before completion with a message that */ * the Hessian is not invertable? The standard textbook advice */ * is to respecify the model, but this is another way of saying */ * that the researcher should change the question being asked. */ * Obviously, however, computer programs should not be in the */ * business of deciding what questions are worthy of */ * study. Although noninvertable Hessians are sometimes signals */ * of poorly posed questions, nonsensical models, or */ * inappropriate estimators, they also frequently occur when */ * information about the quantities of interest exists in the */ * data, through the likelihood function. We explain the */ * problem in some detail and lay out two preliminary proposals */ * for ways of dealing with noninvertable Hessians without */ * changing the question asked. */ ***************************************************************/
- proc gmchol(A);
/* calculates the Gill-Murray generalized choleski decomposition / / input matrix A must be non-singular and symmetric / / Author: Jeff Gill. Part of the Hessian Project. */ local i,j,k,n,sum,R,theta_j,norm_A,phi_j,delta,xi_j,gamm,E,beta_j; n = rows(A); R = eye(n); E = zeros(n,n); norm_A = maxc(sumc(abs(A))); gamm = maxc(abs(diag(A))); delta = maxc(maxc(__macheps*norm_A~__macheps)); for j (1, n, 1);
theta_j = 0; for i (1, n, 1);
sum = 0; for k (1, (i-1), 1);
sum = sum + R[k,i]*R[k,j];endfor; R[i,j] = (A[i,j] - sum)/R[i,i]; if (A[i,j] -sum) > theta_j;
theta_j = A[i,j] - sum;endif; if i > j;
R[i,j] = 0;endif;
endfor; sum = 0; for k (1, (j-1), 1);
sum = sum + R[k,j]^2;endfor; phi_j = A[j,j] - sum; if (j+1) <= n;
xi_j = maxc(abs(A[(j+1):n,j]));- else;
- xi_j = maxc(abs(A[n,j]));
endif; beta_j = sqrt(maxc(maxc(gamm~(xi_j/n)~__macheps))); if delta >= maxc(abs(phi_j)~((theta_j^2)/(beta_j^2)));
E[j,j] = delta - phi_j;- elseif abs(phi_j) >= maxc(((delta^2)/(beta_j^2))~delta);
- E[j,j] = abs(phi_j) - phi_j;
- elseif ((theta_j^2)/(beta_j^2)) >= maxc(delta~abs(phi_j));
- E[j,j] = ((theta_j^2)/(beta_j^2)) - phi_j;
endif; R[j,j] = sqrt(A[j,j] - sum + E[j,j]);
endfor; retp(R’R);
endp;
Examples
>>> np.random.seed(3) >>> A = np.random.rand(100,100)*2 - 1 >>> A = A + A.T >>> L, e = gmchol(A, test=True) >>> np.allclose(np.dot(L,L.T), (A + np.diag(e))) True >>> -e.max()/np.linalg.eigvalsh(A).min() < 1000 True
>>> A2 = np.array([[-0.451, -0.041, 0.124], ... [-0.041, -0.265, 0.061], ... [ 0.124, 0.061, -0.517]]) >>> L, e = gmchol(A2, test=True) >>> e.max()/np.linalg.eigvalsh(A).min()
- mmf.math.linalg.cholesky.gmw81.colmod(A)[source]¶
Modified Cholesky factorization
R = cholmod(A) returns the upper Cholesky factor of A (same as chol) if A is (sufficiently) positive definite, and otherwise returns a modified factor R with diagonal entries >= sqrt(delta) and offdiagonal entries <= beta in absolute value, where delta and beta are defined in terms of size of diagonal and offdiagonal entries of A and the machine precision; see below. The idea is to ensure that E = A - R’*R is reasonably small if A is not too far from being indefinite. If A is sparse, so is R. The output parameter indef is set to 0 if `A is sufficiently positive definite and to 1 if the factorization is modified.
The point of modified Cholesky is to avoid computing eigenvalues, but for dense matrices, MODCHOL takes longer than calling the built-in EIG, because of the cost of interpreting the code, even though it only has one loop and uses vector operations.
reference: Nocedal and Wright, Algorithm 3.4 and subsequent discussion (not Algorithm 3.5, which is more complicated) original algorithm is due to Gill and Murray, 1974 written by M. Overton, overton@cs.nyu.edu, last modified Feb 2005
convenient to work with A = LDL’ where D is diagonal, L is unit lower triangular, and so R = (LD^(1/2))’
Notes
This is based on the following MATLAB code from Michael L. Overton: http://cs.nyu.edu/overton/g22_opt/codes/cholmod.m
function [R, indef, E] = cholmod(A) if sum(sum(abs(A-A'))) > 0 error('A is not symmetric') end % set parameters governing bounds on L and D (eps is machine epsilon) n = length(A); diagA = diag(A); gamma = max(abs(diagA)); % max diagonal entry xi = max(max(abs(A - diag(diagA)))); % max offidagonal entry delta = eps*(max([gamma+xi, 1])); beta = sqrt(max([gamma, xi/n, eps])); indef = 0; % initialize d and L d = zeros(n,1); if issparse(A) L = speye(n); % sparse identity else L = eye(n); end % there are no inner for loops, everything implemented with % vector operations for a reasonable level of efficiency for j = 1:n K = 1:j-1; % column index: all columns to left of diagonal % d(K) doesn't work in case K is empty djtemp = A(j,j) - L(j,K)*(d(K,1).*L(j,K)'); % C(j,j) in book if j < n I = j+1:n; % row index: all rows below diagonal Ccol = A(I,j) - L(I,K)*(d(K,1).*L(j,K)'); % C(I,j) in book theta = max(abs(Ccol)); % guarantees d(j) not too small and L(I,j) not too big % in sufficiently positive definite case, d(j) = djtemp d(j) = max([abs(djtemp), (theta/beta)^2, delta]); L(I,j) = Ccol/d(j); else d(j) = max([abs(djtemp), delta]); end if d(j) > djtemp % A was not sufficiently positive definite indef = 1; end end % convert to usual output format: replace L by L*sqrt(D) and transpose for j=1:n L(:,j) = L(:,j)*sqrt(d(j)); % L = L*diag(sqrt(d)) bad in sparse case end; R = L'; if nargout == 3 E = A - R'*R; endExamples
>>> np.random.seed(3) >>> A = np.random.rand(100,100)*2-1 >>> A = A + A.T >>> L, e = colmod(A) >>> np.allclose(np.dot(L, L.T) - np.diag(e), A) True >>> 0 < np.linalg.eigvalsh(A + np.diag(e)).min() True >>> -e.max()/np.linalg.eigvalsh(A).min() < 1000 True
- mmf.math.linalg.cholesky.gmw81.mchol2(A)[source]¶
- Based on the MATLAB code
by Brian Borchers and Michael Zibulevsky:
% % [L,D,E,pneg]=mchol2(G) % % Given a symmetric matrix G, find a matrix E of "small" norm and c % L, and D such that G+E is Positive Definite, and % % G+E = L*D*L' % % Also, calculate a direction pneg, such that if G is not PSD, then % % pneg'*G*pneg < 0 % % Reference: Gill, Murray, and Wright, "Practical Optimization", p111. % Author: Brian Borchers (borchers@nmt.edu) % Modification: Michael Zibulevsky (mzib@ee.technion.ac.il) % function [L,D,E,pneg]=mcholmz3(G) % % n gives the size of the matrix. % n=size(G,1); % % gamma, zi, nu, and beta2 are quantities used by the algorithm. % gamma=max(diag(G)); zi=max(max(G-diag(diag(G)))); nu=max([1,sqrt(n^2-1)]); beta2=max([gamma, zi/nu, 1.0E-15]); C=diag(diag(G)); L=zeros(n); D=zeros(n,1); % use vestors as diagonal matrices E=zeros(n,1); %%****************** ************** % % Loop through, calculating column j of L for j=1:n % for j=1:n, bb=[1:j-1]; ee=[j+1:n]; if (j > 1), L(j,bb)=C(j,bb)./D(bb)'; % Calculate the jth row of L. end; if (j >= 2) if (j < n), C(ee,j)=G(ee,j)- C(ee,bb)*L(j,bb)'; % Update the jth column of C. end; else C(ee,j)=G(ee,j); end; %%%% Update theta. if (j == n) theta(j)=0; else theta(j)=max(abs(C(ee,j))); end; %%%%% Update D D(j)=max([eps,abs(C(j,j)),theta(j)^2/beta2]'); %%%%%% Update E. E(j)=D(j)-C(j,j); %%%%%%% Update C again... %for i=j+1:n, % C(i,i)=C(i,i)-C(i,j)^2/D(j,j); %end; ind=[j*(n+1)+1 : n+1 : n*n]'; C(ind)=C(ind)-(1/D(j))*C(ee,j).^2; end; % % Put 1's on the diagonal of L % %for j=1:n, % L(j,j)=1; %end; ind=[1 : n+1 : n*n]'; L(ind)=1; % % if needed, finded a descent direction. % if (nargout == 4) [m,col]=min(diag(C)); rhs=zeros(n,1); rhs(col)=1; pneg=L'- hs;
end;
return
%%%%%%%%%%%%%%%%%%%%%% Test %%%%%%%%%%%%%
n=3; G=rand(n);G=G+G’;eigG=eig(G), [L,D,E,pneg]=mchol(G) [L1,D1,E1,pneg1]=mcholmz1(G); LL=L-L1, DD=D-D1, EE=E-E1, pp=pneg-pneg1
n=3; G=rand(n);G=G+G’;eigG=eig(G), [L,D,E,pneg]=mchol(G) [L2,D2,E2,pneg2]=mcholmz2(G); LL=L-L2, DD=D-diag(D2), EE=E-diag(E2), pp=pneg-pneg2