8 Week 8
Topic: Add Health data: variable creation and scale scoring
This week's lesson will provide more background on variable creation and scale scoring. The scale scoring exercise will be used to create a single variable that represents how well respondents did overall on a subset of questions.
8.1 Scale scoring
We will be using data from the Knowledge Quiz. Download or open 21600-0001-Codebook_Questionnaire.pdf in a new window or tab and go to page 203, or search for the string H1KQ1A.
We will be using the file AHwave1_v1.dta, which is downloaded and read in the following code chunk, along with presentation of the column names, labels, and values.
dat <- haven::read_dta("http://staff.washington.edu/phurvitz/csde502_winter_2021/data/AHwave1_v1.dta")
metadata <- bind_cols(
# variable name
varname = colnames(dat),
# label
varlabel = lapply(dat, function(x) attributes(x)$label) %>%
unlist(),
# values
varvalues = lapply(dat, function(x) attributes(x)$labels) %>%
# names the variable label vector
lapply(., function(x) names(x)) %>%
# as character
as.character() %>%
# remove the c() construction
str_remove_all("^c\\(|\\)$")
)
DT::datatable(metadata)Questions H1KQ1A, H1KQ2A, ..., H1KQ10A are factual questions about contraception that are administered to participants \(\ge\) age 15. We will be creating a single score that sums up all the correct answers across these questions for each participant \(\ge\) age 15. Because the set of questions is paired, with question "a" being the factual portion and "b" being the level of confidence, we want only those questions with column names ending with "a".
8.1.1 Selecting specific columns
There are several ways of selecting the desired columns into a new data frame. Here is brute force approach:
# create a data frame of some columns and age >= 15
mydat_bruteforce <- dat %>%
# drop those under 15 y
filter(h1kq1a != 7) %>%
# get answers
select(
aid, # subject ID
h1kq1a,
h1kq2a,
h1kq3a,
h1kq4a,
h1kq5a,
h1kq6a,
h1kq7a,
h1kq8a,
h1kq9a,
h1kq10a
)Although there were only 10 columns with this name pattern, what if there had been 30 or 50? You would not want to have to enter each column name separately. Not only would this be tedious, there would always be the possibility of making a keyboarding mistake.
Instead of the brute force approach, we can use the matches() function with a regular expression. The regular expression here is ^h1kq.*a$, which translates to "at the start of the string, match h1kq, then any number of any characters, then a followed by the end of the string".
mydat <- dat %>%
filter(h1kq1a != 7) %>%
select(
aid,
matches("h1kq.*a")
)We check that both processes yielded the same result:
identical(mydat_bruteforce, mydat)## [1] TRUE8.1.2 Comparing participant answers to correct answers
Now that we have a data frame limited to the participants in the correct age range and only the questions we want, we need to set up tests for whether the questions were answered correctly or not. From the metadata we can see that for some questions, the correct answer was (1) true and for some, the correct answer was (2) false.
We can scan through the questions in the metadata to create a vector of correct answers:
# the correct answers
correct <- c(2, 1, 2, 2, 2, 2, 2, 1, 2, 2)
# make a named vector of the answers using the selected column names
names(correct) <- str_subset(string = names(mydat),
pattern = "h1kq.*a")What we now need to do is compare this vector to a vector constructed of the answers in mydat. There are a few approaches that could be taken. A brute force approach could use a loop to iterate over each record in the answers, and for each record to iterate over each answer:
# time this
t0 <- Sys.time()
# make an output
ans_loop <- NULL
# iterate over rows
#testing:
#for(i in 1:3){
for(i in 1:nrow(mydat)){
# init a vector
Q <- NULL
# iterate over columns, ignoring the first "aid" column
for(j in 2:ncol(mydat)){
# get the value of the answer
ans_subj <- mydat[i, j]
# get the correct answer
ans_actual <- correct[j - 1]
# compare
cmp <- ans_actual == ans_subj
# append
Q <- c(Q, cmp)
}
# append
ans_loop <- rbind(ans_loop, Q)
}
# package it up nicely
ans_loop %<>% data.frame()
colnames(ans_loop) <- names(correct)
row.names(ans_loop) <- NULL
# timing
t1 <- Sys.time()
runtime_loop <- difftime(t1, t0, units = "secs") %>% as.numeric() %>% round(1)It took 44.2 s to run. This low performance is because the algorithm is visiting every cell and comparing one-by-on with a rotating value for the correct answer from the vector of correct answers. Each object is required to be handled separately in RAM as the process continues.
Another approach uses plyr::adply(), which runs a function over a set of rows. The plyr package contains a set of tools for splitting data, applying functions, and recombining.
# time this
t0 <- Sys.time()
ans_adply <- mydat %>%
select(-1) %>%
plyr::adply(.margins = 1,
function(x) x == correct)
t1 <- Sys.time()
runtime_adply <- difftime(t1, t0, units = "secs") %>% as.numeric() %>% round(1)The adply() version takes far less coding, but still took 41 s to run.
Yet another different approach compares the data frame of participant answers to the vector of correct answers. The correct answers vector will get recycled until all values have been processed. The problem with this method is that the comparison runs down columns rather than across rows.
This demonstrates the problem. Table 8.1 shows a pattern of "correct" values, and Table 8.2 shows a table of responses
# make a pattern to match against
pat1 <- c(1, 2, 3, 4)
names(pat1) <- paste("question", 1:4, sep="_")
pat1 %>%
t() %>%
data.frame() %>%
kable(caption = 'A pattern of "correct" values') %>%
kable_styling(full_width = FALSE, position = "left")| question_1 | question_2 | question_3 | question_4 |
|---|---|---|---|
| 1 | 2 | 3 | 4 |
# make a data frame to process
d1 <- cbind(rep(1, 3), rep(2, 3), rep(3,3), rep(4, 3)) %>%
data.frame()
names(d1) <- names(pat1)
d1 %>%
kable(caption = "A table of responses") %>%
kable_styling(full_width = FALSE, position = "left")| question_1 | question_2 | question_3 | question_4 |
|---|---|---|---|
| 1 | 2 | 3 | 4 |
| 1 | 2 | 3 | 4 |
| 1 | 2 | 3 | 4 |
The pattern matches the first row of data (Table 8.3)
(pat1 == d1[1,]) %>%
kable(caption = "Matches for the first row") %>%
kable_styling(full_width = FALSE, position = "left") | question_1 | question_2 | question_3 | question_4 |
|---|---|---|---|
| TRUE | TRUE | TRUE | TRUE |
The patterns do not match the overall table as might be expected (??).
(d1 == pat1) %>%
kable() %>%
kable_styling(full_width = FALSE, position = "left")| question_1 | question_2 | question_3 | question_4 |
|---|---|---|---|
| TRUE | FALSE | TRUE | FALSE |
| FALSE | FALSE | FALSE | FALSE |
| FALSE | TRUE | FALSE | TRUE |
In order to match the pattern to each row, a transpose is required. The following code performs the transpose, pattern match, and re-transpose, with results in 8.4
# transpose, check for matching and transpose back
(d1 %>% t() == pat1) %>%
t() %>%
kable(caption = "Expected pattern matches") %>%
kable_styling(full_width = FALSE, position = "left") | question_1 | question_2 | question_3 | question_4 |
|---|---|---|---|
| TRUE | TRUE | TRUE | TRUE |
| TRUE | TRUE | TRUE | TRUE |
| TRUE | TRUE | TRUE | TRUE |
So the trick is to use a transpose (t()) to swap rows and columns. Then unlist() will enforce the correct ordering. After running the comparison, the data are transposed again to recreate the original structure.
# time this
t0 <- Sys.time()
# transpose and compare
ans_unlist <- mydat %>%
select(-1) %>%
t(.) %>%
unlist(.) == correct
# re-transpose and make a data frame
ans_unlist %<>%
t(.) %>%
data.frame()
# column names
colnames(ans_unlist) <- names(correct)
t1 <- Sys.time()
runtime_unlist <- difftime(t1, t0, units = "secs") %>% as.numeric() %>% round(2)This method took 0.01 s to complete.
Yet another method similarly uses the double transpose method.
# time this
t0 <- Sys.time()
# strip the ID column and transpose
z <- mydat %>%
select(-1) %>%
t()
# compare, transpose, and make a data frame
ans_tranpose <- (z == correct) %>%
t(.) %>%
data.frame()
t1 <- Sys.time()
runtime_transpose <- difftime(t1, t0, units = "secs") %>% as.numeric() %>% round(2)This method took 0.01 s to complete.
The final and most direct method uses base::sweep(), which can be used to compare a vector against all rows or columns in a data frame. In order to use this function, the data frame needs to have the same number of rows (or columns) as the comparison vector. So any additional rows or columns need to be stripped. Because we may have additional columns (e.g., aid), those must be removed before running sweep(), then added back in again. Additionally, the result of sweep() is a matrix, so it needs to be converted to a data frame for greater functionality.
t0 <- Sys.time()
ans_sweep <- mydat %>%
# drop the aid column
select(-aid) %>%
# run the wweep
sweep(x = ., MARGIN = 2, STATS = correct, FUN = "==") %>%
# convert to data frame
data.frame()
t1 <- Sys.time()
runtime_sweep <- difftime(t1, t0, units = "secs") %>% as.numeric() %>% round(3)The sweep() method took 0.019 s.
We should check that the methods all gave identical answers.
Vectorize(identical, 'x')(list(ans_loop, ans_adply, ans_tranpose, ans_unlist), ans_loop)## [1] TRUE TRUE TRUE TRUE8.1.3 Scoring across columns
Now that we have a data frame indicating for each participant whether they answered each question correctly, we can total the number of correct answers for each participant. The rowSums() function allows sums across rows. Because the logical values are automatically converted to numerical values (TRUE = 1; FALSE = 0), the sums provide the total number of correct answers per participant. Also because the data frame only consists of answers 1 .. 10, we can use an unqualified rowSums(), otherwise it would be necessary to specify which columns would be included by either position or column name.
We also bring the subject identifier (aid) back in and reorder the columns with select(). Note that after the specified aid and total h1kqNa_sum columns, we can use everything() to select the remainder of the columns.
ans_loop %<>%
# calculate the rowSums
mutate(h1kqNa_sum = rowSums(.)) %>%
# bring the ID back in
mutate(aid = mydat$aid) %>%
# reorder columns
select(aid, h1kqNa_sum, everything())To show differences in total score by sex, we can join the main data back using the aid identifier and create a simple graph. Figure 8.1 shows that more females than males had overall higher counts of correct scores on the Knowledge Quiz.
ans_loop %<>%
left_join(dat, by = "aid") %>%
mutate(
sex = case_when(
bio_sex == 1 ~ 'male',
bio_sex == 2 ~ 'female'
)
)
ggplot(data = ans_loop, mapping = aes(x = h1kqNa_sum))+
geom_histogram(stat = "count") +
facet_grid(sex ~ .) +
xlab("correct answers on Knowledge Quiz") +
scale_x_continuous(breaks=0:10)## Warning: Ignoring unknown parameters: binwidth, bins, pad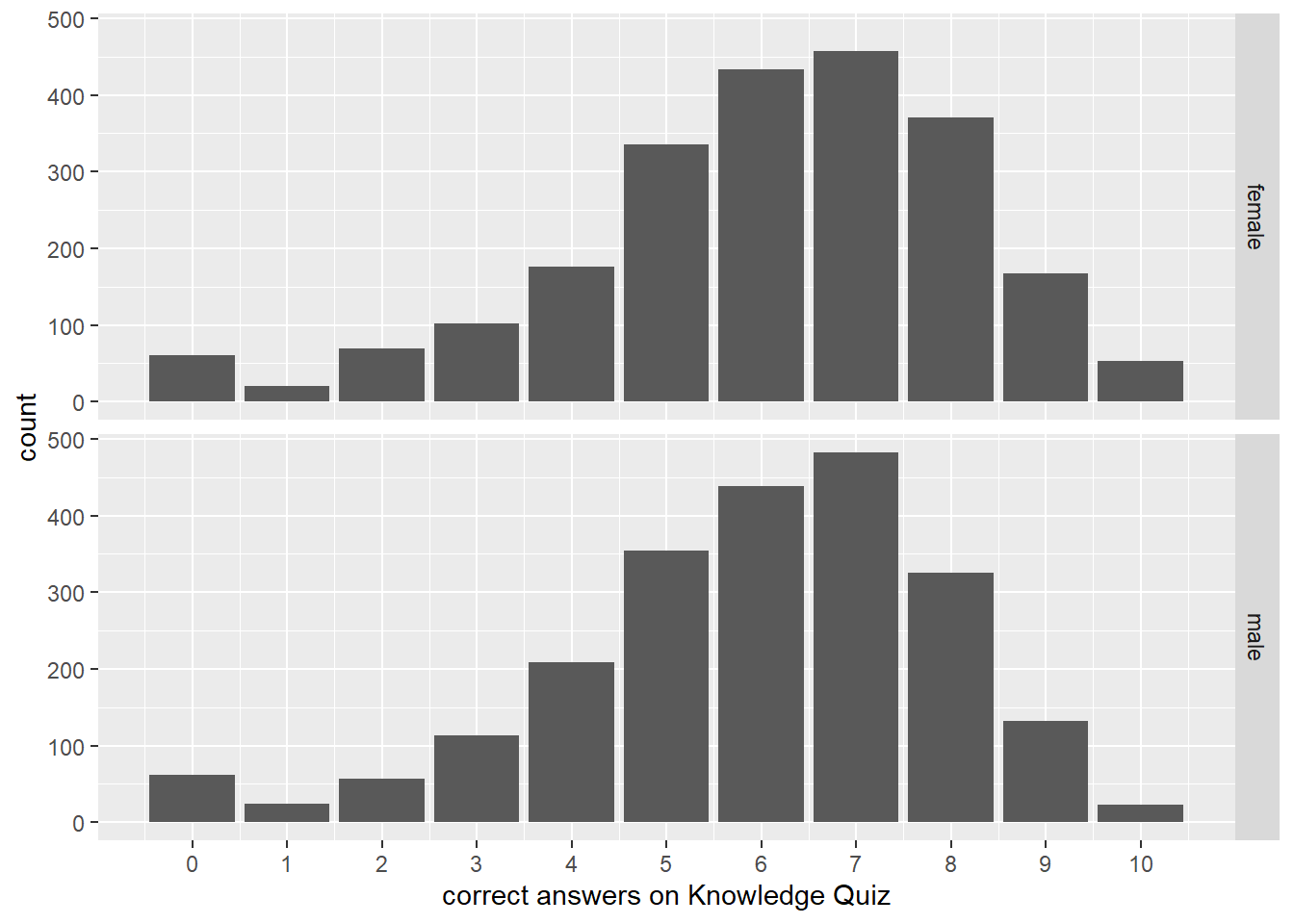
Figure 8.1: Histogram of count of correct answers on Knowledge Quiz stratified by sex of respondent
8.2 Reordering values
Sometimes variables are provided in the reverse order of what you might want. For example, the answers pertaining to confidence in the Knowledge Quiz are in this specific order:
attributes(dat$h1kq1b)$labels %>% t() %>% t() %>% data.frame()## .
## (1) Very 1
## (2) Moderately 2
## (3) Slightly 3
## (4) Not at all 4
## (6) Refused 6
## (7) Legitimate skip (less than 15) 7
## (8) Don't know 8
## (9) Not applicable 9To come up with a scale score for these, it would be better to have Very valued as a 4 and Not at all as a 1 so that row-wise sums would yield higher values for those who were more confident in many answers. One could use the existing values, but then the interpretation of an overall confidence score might be difficult, with the most confidence for the lowest overall score.
Changing these values is quite straightforward. The case_when() function can be used:
# for comparison, make a backup data frame
datbak <- dat2 <- dat
# reassign values
dat %<>%
mutate(h1kq1b =
case_when(
# main changes
h1kq1b == 4 ~ 1,
h1kq1b == 3 ~ 2,
h1kq1b == 2 ~ 3,
h1kq1b == 1 ~ 4,
# anythng that is not in the above list gets its original value
TRUE ~ as.numeric(h1kq1b))
)It is a bit more awkward to deal with multiple columns. One might be tempted to use a brute force method by copy/paste/edit but using the mutate_at() function can help through the use of regular expression pattern matching for column names. The same function will be performed on multiple columns. Here we use a similar regular expression to find the columns representing confidence in answers to the Knowledge Quiz (h1kq.*b). The use of the dot (.) is shorthand for "the current object" which in this case is the specified column.
dat2 %<>%
mutate_at(.vars = vars(matches("h1kq.*b")),
funs(
case_when(
. == 4 ~ 1,
. == 3 ~ 2,
. == 2 ~ 3,
. == 1 ~ 4,
TRUE ~ as.numeric(.)
)
)
)For the sake of comparison to show that the single bit of code acted on multiple columns:
orig1 <- table(datbak$h1kq1b) %>% data.frame()
mod1 <- table(dat2$h1kq1b) %>% data.frame()
orig2 <- table(datbak$h1kq2b) %>% data.frame()
mod2 <- table(dat2$h1kq2b) %>% data.frame()
cbind(orig1, mod1, orig2, mod2) %>%
kable() %>%
kable_styling(full_width = FALSE, position = "left") %>%
add_header_above(rep(c("original" = 2, "modified" = 2), 2)) %>%
add_header_above(c("h1kq1b" = 4, "h1kq2b" = 4))| Var1 | Freq | Var1 | Freq | Var1 | Freq | Var1 | Freq |
|---|---|---|---|---|---|---|---|
| 1 | 1070 | 1 | 373 | 1 | 1658 | 1 | 289 |
| 2 | 1723 | 2 | 1109 | 2 | 1542 | 2 | 800 |
| 3 | 1109 | 3 | 1723 | 3 | 800 | 3 | 1542 |
| 4 | 373 | 4 | 1070 | 4 | 289 | 4 | 1658 |
| 6 | 27 | 6 | 27 | 6 | 32 | 6 | 32 |
| 7 | 2039 | 7 | 2039 | 7 | 2039 | 7 | 2039 |
| 8 | 162 | 8 | 162 | 8 | 143 | 8 | 143 |
| 9 | 1 | 9 | 1 | 9 | 1 | 9 | 1 |
# too many unique combinations!
# dat2 %>%
# group_by_at(vars(matches("h1kq.*b"))) %>%
# dplyr::summarise(n = n())Now that the values are reordered, they can be used in multiple-column scale scoring as demonstrated above.
8.3 Source code
cat(readLines(con = "08-week08.Rmd"), sep = '\n')# Week 8 {#week8}
```{r, echo=FALSE, warning=FALSE, message=FALSE}
library(tidyverse)
library(magrittr)
library(knitr)
library(kableExtra)
library(haven)
library(pdftools)
library(curl)
library(ggplot2)
```
<h2>Topic: Add Health data: variable creation and scale scoring</h2>
This week's lesson will provide more background on variable creation and scale scoring. The scale scoring exercise will be used to create a single variable that represents how well respondents did overall on a subset of questions.
## Scale scoring
We will be using data from the Knowledge Quiz. Download or open [21600-0001-Codebook_Questionnaire.pdf](http://staff.washington.edu/phurvitz/csde502_winter_2021/data/metadata/Wave1_Comprehensive_Codebook/21600-0001-Codebook_Questionnaire.pdf) in a new window or tab and go to page 203, or search for the string `H1KQ1A`.
We will be using the file `AHwave1_v1.dta`, which is downloaded and read in the following code chunk, along with presentation of the column names, labels, and values.
```{r}
dat <- haven::read_dta("http://staff.washington.edu/phurvitz/csde502_winter_2021/data/AHwave1_v1.dta")
metadata <- bind_cols(
# variable name
varname = colnames(dat),
# label
varlabel = lapply(dat, function(x) attributes(x)$label) %>%
unlist(),
# values
varvalues = lapply(dat, function(x) attributes(x)$labels) %>%
# names the variable label vector
lapply(., function(x) names(x)) %>%
# as character
as.character() %>%
# remove the c() construction
str_remove_all("^c\\(|\\)$")
)
DT::datatable(metadata)
```
Questions `H1KQ1A`, `H1KQ2A`, ..., `H1KQ10A` are factual questions about contraception that are administered to participants $\ge$ age 15. We will be creating a single score that sums up all the correct answers across these questions for each participant $\ge$ age 15. Because the set of questions is paired, with question "a" being the factual portion and "b" being the level of confidence, we want only those questions with column names ending with "a".
### Selecting specific columns
There are several ways of selecting the desired columns into a new data frame. Here is brute force approach:
```{r}
# create a data frame of some columns and age >= 15
mydat_bruteforce <- dat %>%
# drop those under 15 y
filter(h1kq1a != 7) %>%
# get answers
select(
aid, # subject ID
h1kq1a,
h1kq2a,
h1kq3a,
h1kq4a,
h1kq5a,
h1kq6a,
h1kq7a,
h1kq8a,
h1kq9a,
h1kq10a
)
```
Although there were only 10 columns with this name pattern, what if there had been 30 or 50? You would not want to have to enter each column name separately. Not only would this be tedious, there would always be the possibility of making a keyboarding mistake.
Instead of the brute force approach, we can use the `matches()` function with a regular expression. The regular expression here is `^h1kq.*a$`, which translates to "at the start of the string, match `h1kq`, then any number of any characters, then `a` followed by the end of the string".
```{r}
mydat <- dat %>%
filter(h1kq1a != 7) %>%
select(
aid,
matches("h1kq.*a")
)
```
We check that both processes yielded the same result:
```{r}
identical(mydat_bruteforce, mydat)
```
### Comparing participant answers to correct answers
Now that we have a data frame limited to the participants in the correct age range and only the questions we want, we need to set up tests for whether the questions were answered correctly or not. From the metadata we can see that for some questions, the correct answer was `(1) true` and for some, the correct answer was `(2) false`.
We can scan through the questions in the metadata to create a vector of correct answers:
```{r}
# the correct answers
correct <- c(2, 1, 2, 2, 2, 2, 2, 1, 2, 2)
# make a named vector of the answers using the selected column names
names(correct) <- str_subset(string = names(mydat),
pattern = "h1kq.*a")
```
What we now need to do is compare this vector to a vector constructed of the answers in `mydat`. There are a few approaches that could be taken. A brute force approach could use a loop to iterate over each record in the answers, and for each record to iterate over each answer:
```{r}
# time this
t0 <- Sys.time()
# make an output
ans_loop <- NULL
# iterate over rows
#testing:
#for(i in 1:3){
for(i in 1:nrow(mydat)){
# init a vector
Q <- NULL
# iterate over columns, ignoring the first "aid" column
for(j in 2:ncol(mydat)){
# get the value of the answer
ans_subj <- mydat[i, j]
# get the correct answer
ans_actual <- correct[j - 1]
# compare
cmp <- ans_actual == ans_subj
# append
Q <- c(Q, cmp)
}
# append
ans_loop <- rbind(ans_loop, Q)
}
# package it up nicely
ans_loop %<>% data.frame()
colnames(ans_loop) <- names(correct)
row.names(ans_loop) <- NULL
# timing
t1 <- Sys.time()
runtime_loop <- difftime(t1, t0, units = "secs") %>% as.numeric() %>% round(1)
```
It took `r runtime_loop` s to run. This low performance is because the algorithm is visiting every cell and comparing one-by-on with a rotating value for the correct answer from the vector of correct answers. Each object is required to be handled separately in RAM as the process continues.
Another approach uses `plyr::adply()`, which runs a function over a set of rows. The [`plyr`](https://www.rdocumentation.org/packages/plyr/) package contains a set of tools for splitting data, applying functions, and recombining.
```{r}
# time this
t0 <- Sys.time()
ans_adply <- mydat %>%
select(-1) %>%
plyr::adply(.margins = 1,
function(x) x == correct)
t1 <- Sys.time()
runtime_adply <- difftime(t1, t0, units = "secs") %>% as.numeric() %>% round(1)
```
The `adply()` version takes far less coding, but still took `r runtime_adply` s to run.
Yet another different approach compares the data frame of participant answers to the vector of correct answers. The correct answers vector will get recycled until all values have been processed. The problem with this method is that the comparison runs down columns rather than across rows.
This demonstrates the problem. Table \@ref(tab:pat1vector) shows a pattern of "correct" values, and Table \@ref(tab:d1dataframe) shows a table of responses
```{r pat1vector}
# make a pattern to match against
pat1 <- c(1, 2, 3, 4)
names(pat1) <- paste("question", 1:4, sep="_")
pat1 %>%
t() %>%
data.frame() %>%
kable(caption = 'A pattern of "correct" values') %>%
kable_styling(full_width = FALSE, position = "left")
```
```{r d1dataframe}
# make a data frame to process
d1 <- cbind(rep(1, 3), rep(2, 3), rep(3,3), rep(4, 3)) %>%
data.frame()
names(d1) <- names(pat1)
d1 %>%
kable(caption = "A table of responses") %>%
kable_styling(full_width = FALSE, position = "left")
```
The pattern matches the first row of data (Table \@ref(tab:patmatchonerow))
```{r patmatchonerow}
(pat1 == d1[1,]) %>%
kable(caption = "Matches for the first row") %>%
kable_styling(full_width = FALSE, position = "left")
```
The patterns do not match the overall table as might be expected (\@ref(tab:patmatchdfbad)).
```{r patmatchdfbad, fig.cap="Unexpected pattern matches"}
(d1 == pat1) %>%
kable() %>%
kable_styling(full_width = FALSE, position = "left")
```
In order to match the pattern to each row, a transpose is required. The following code performs the transpose, pattern match, and re-transpose, with results in \@ref(tab:patmatchdfgood)
```{r patmatchdfgood}
# transpose, check for matching and transpose back
(d1 %>% t() == pat1) %>%
t() %>%
kable(caption = "Expected pattern matches") %>%
kable_styling(full_width = FALSE, position = "left")
```
So the trick is to use a transpose (`t()`) to swap rows and columns. Then `unlist()` will enforce the correct ordering. After running the comparison, the data are transposed again to recreate the original structure.
```{r}
# time this
t0 <- Sys.time()
# transpose and compare
ans_unlist <- mydat %>%
select(-1) %>%
t(.) %>%
unlist(.) == correct
# re-transpose and make a data frame
ans_unlist %<>%
t(.) %>%
data.frame()
# column names
colnames(ans_unlist) <- names(correct)
t1 <- Sys.time()
runtime_unlist <- difftime(t1, t0, units = "secs") %>% as.numeric() %>% round(2)
```
This method took `r runtime_unlist` s to complete.
Yet another method similarly uses the double transpose method.
```{r}
# time this
t0 <- Sys.time()
# strip the ID column and transpose
z <- mydat %>%
select(-1) %>%
t()
# compare, transpose, and make a data frame
ans_tranpose <- (z == correct) %>%
t(.) %>%
data.frame()
t1 <- Sys.time()
runtime_transpose <- difftime(t1, t0, units = "secs") %>% as.numeric() %>% round(2)
```
This method took `r runtime_transpose` s to complete.
The final and most direct method uses `base::sweep()`, which can be used to compare a vector against all rows or columns in a data frame. In order to use this function, the data frame needs to have the same number of rows (or columns) as the comparison vector. So any additional rows or columns need to be stripped. Because we may have additional columns (e.g., `aid`), those must be removed before running `sweep()`, then added back in again. Additionally, the result of `sweep()` is a matrix, so it needs to be converted to a data frame for greater functionality.
```{r}
t0 <- Sys.time()
ans_sweep <- mydat %>%
# drop the aid column
select(-aid) %>%
# run the wweep
sweep(x = ., MARGIN = 2, STATS = correct, FUN = "==") %>%
# convert to data frame
data.frame()
t1 <- Sys.time()
runtime_sweep <- difftime(t1, t0, units = "secs") %>% as.numeric() %>% round(3)
```
The `sweep()` method took `r runtime_sweep` s.
We should check that the methods all gave identical answers.
```{r}
Vectorize(identical, 'x')(list(ans_loop, ans_adply, ans_tranpose, ans_unlist), ans_loop)
```
### Scoring across columns{#scoring-across-columns}
Now that we have a data frame indicating for each participant whether they answered each question correctly, we can total the number of correct answers for each participant. The `rowSums()` function allows sums across rows. Because the logical values are automatically converted to numerical values (TRUE = 1; FALSE = 0), the sums provide the total number of correct answers per participant. Also because the data frame only consists of answers 1 .. 10, we can use an unqualified `rowSums()`, otherwise it would be necessary to specify which columns would be included by either position or column name.
We also bring the subject identifier (`aid`) back in and reorder the columns with `select()`. Note that after the specified `aid` and total `h1kqNa_sum` columns, we can use `everything()` to select the remainder of the columns.
```{r}
ans_loop %<>%
# calculate the rowSums
mutate(h1kqNa_sum = rowSums(.)) %>%
# bring the ID back in
mutate(aid = mydat$aid) %>%
# reorder columns
select(aid, h1kqNa_sum, everything())
```
To show differences in total score by sex, we can join the main data back using the `aid` identifier and create a simple graph. Figure \@ref(fig:hist) shows that more females than males had overall higher counts of correct scores on the Knowledge Quiz.
```{r hist, fig.cap="Histogram of count of correct answers on Knowledge Quiz stratified by sex of respondent"}
ans_loop %<>%
left_join(dat, by = "aid") %>%
mutate(
sex = case_when(
bio_sex == 1 ~ 'male',
bio_sex == 2 ~ 'female'
)
)
ggplot(data = ans_loop, mapping = aes(x = h1kqNa_sum))+
geom_histogram(stat = "count") +
facet_grid(sex ~ .) +
xlab("correct answers on Knowledge Quiz") +
scale_x_continuous(breaks=0:10)
```
## Reordering values
Sometimes variables are provided in the reverse order of what you might want. For example, the answers pertaining to confidence in the Knowledge Quiz are in this specific order:
```{r}
attributes(dat$h1kq1b)$labels %>% t() %>% t() %>% data.frame()
```
To come up with a scale score for these, it would be better to have `Very` valued as a `4` and `Not at all` as a `1` so that row-wise sums would yield higher values for those who were more confident in many answers. One could use the existing values, but then the interpretation of an overall confidence score might be difficult, with the most confidence for the lowest overall score.
Changing these values is quite straightforward. The `case_when()` function can be used:
```{r}
# for comparison, make a backup data frame
datbak <- dat2 <- dat
# reassign values
dat %<>%
mutate(h1kq1b =
case_when(
# main changes
h1kq1b == 4 ~ 1,
h1kq1b == 3 ~ 2,
h1kq1b == 2 ~ 3,
h1kq1b == 1 ~ 4,
# anythng that is not in the above list gets its original value
TRUE ~ as.numeric(h1kq1b))
)
```
It is a bit more awkward to deal with multiple columns. One might be tempted to use a brute force method by copy/paste/edit but using the `mutate_at()` function can help through the use of regular expression pattern matching for column names. The same function will be performed on multiple columns. Here we use a similar regular expression to find the columns representing confidence in answers to the Knowledge Quiz (`h1kq.*b`). The use of the dot (`.`) is shorthand for "the current object" which in this case is the specified column.
```{r, warning=FALSE}
dat2 %<>%
mutate_at(.vars = vars(matches("h1kq.*b")),
funs(
case_when(
. == 4 ~ 1,
. == 3 ~ 2,
. == 2 ~ 3,
. == 1 ~ 4,
TRUE ~ as.numeric(.)
)
)
)
```
For the sake of comparison to show that the single bit of code acted on multiple columns:
```{r}
orig1 <- table(datbak$h1kq1b) %>% data.frame()
mod1 <- table(dat2$h1kq1b) %>% data.frame()
orig2 <- table(datbak$h1kq2b) %>% data.frame()
mod2 <- table(dat2$h1kq2b) %>% data.frame()
cbind(orig1, mod1, orig2, mod2) %>%
kable() %>%
kable_styling(full_width = FALSE, position = "left") %>%
add_header_above(rep(c("original" = 2, "modified" = 2), 2)) %>%
add_header_above(c("h1kq1b" = 4, "h1kq2b" = 4))
# too many unique combinations!
# dat2 %>%
# group_by_at(vars(matches("h1kq.*b"))) %>%
# dplyr::summarise(n = n())
```
Now that the values are reordered, they can be used in multiple-column scale scoring as demonstrated above.
## Source code
[08-week08.Rmd](08-week08.Rmd)
```{r comment=''}
cat(readLines(con = "08-week08.Rmd"), sep = '\n')
```