 Plant Twitter : Ligands Under 140 Amino Acids Enforcing Stomatal Patterning Plant Twitter : Ligands Under 140 Amino Acids Enforcing Stomatal Patterning
Stomata are an essential land plant innovation whose patterning and density are under genetic and environmental control. Recently, several putative ligands have been discovered that influence stomatal density, and they all belong to the EPIDERMAL PATTERNING FACTOR-LIKE family of secreted cysteine-rich peptides. Two of these putative ligands, EPF1 and EPF2, are expressed exclusively in the stomatal lineage cells and negatively regulate stomatal density. A third, EPFL6 or CHALLAH, is also a negative regulator of density, but is expressed subepidermally in the hypocotyl. A fourth, EPFL9 or STOMAGEN, is expressed in the mesophyll tissues and is a positive regulator of density. Genetic evidence suggests that these ligands may compete for the same receptor complex. Proper stomatal patterning is likely to be an intricate process involving ligand competition, regional specificity, and communication between tissue layers. EPFL-family genes exist in the moss Physcomitrella patens, the lycophyte Selaginella moellendorffii, and rice, Oryza sativa, and their sequence analysis yields several genes some of which are related to EPF1, EPF2, EPFL6, and EPFL9. Presence of these EPFL family members in the basal land plants suggests an exciting hypothesis that the genetic components for stomatal patterning originated early in land plant evolution.
In making the evolutionary transition from water to land, one of the greatest challenges for plants was to prevent desiccation while allowing for gas exchange. A major innovation in the form of stomata helped solve this key problem. Stomata likely evolved just once in the ancestor to land plants and are only present on the sporophyte (Raven 2002). Stomata function through adjustment of their aperture by guard cells, which close with loss of turgor pressure and open by drawing in water from neighboring cells. To function properly, then, it is important that stomata not be adjacent to one another, and indeed during development stomata are nearly always formed with at least one non-stomatal cell between them, the so-called one-cell spacing rule (Sachs 1991). While environmental cues, such as light and CO2 concentration, may in part regulate the density of stomata, a genetic mechanism enforces the one-cell spacing rule (Casson and Gray, 2008). Even before the molecular basis behind this rule was known, it was hypothesized that inhibitory cell-cell signals from developing stomatal-lineage cells prevent the development of stomatal cell state in their neighbors (Sachs 1991).
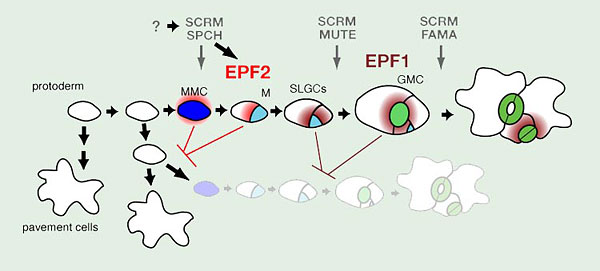
Fig. 1. Stomatal development in Arabidopsis
Three paralogous bHLH transcription factors, SPEECHLESS (SPCH), MUTE, and FAMA, control sequential cell-state transitions during stomatal development. They heterodimerize with unrelated bHLHs SCREAM (SCRM) and SCRM2, which redundantly coordinate their action. SPCH directs the initial entry into the stomatal cell lineage, in which a protodermal cell becomes a meristemoid mother cell (MMC, blue). Non-MMC protodermal cells differentiate into epidermal pavement cells. The MMC divides asymmetrically one or more times, forming a meristemoid (M, cyan) and a stomatal lineage ground cell (SLGC, white), which could be regulated by SPCH as well. On completion of asymmetric divisions, the M becomes a guard mother cell (GMC, light green) in a step controlled by MUTE. The final stoma is formed by a single symmetric division on the GMC into two guard cells (GC, green). FAMA limits this division of the GMC and allows terminal differentiation of the GC to form the stomatal pore. Early stomatal lineage cells secrete EPIDERMAL PATTERNING FACTOR 2 (EPF2, red), a putative ligand which blocks the protoderm to MMC transition, while later stomatal lineage cells produce EPF1 (dark red), which inhibits stomatal lineage identity after entry divisions have occurred.
The process of stomatal development in the model dicot, Arabidopsis thaliana, takes place over several discrete steps, each with a characteristic stomatal lineage cell type (Fig. 1). The first step in stomatal development is entry of an undifferentiated protodermal cell into the stomatal lineage by adopting meristemoid mother cell (MMC) state and commencing asymmetric cell divisions. This entry into asymmetric cell divisions is controlled by the basic helix-loop-helix (bHLH) transcription factor SPEECHLESS (SPCH)(Fig. 1). spch mutants develop with an epidermis composed entirely of non-stomatal pavement cells (MacAlister et al. 2007; Pillitteri et al. 2007; Pillitteri and Torii 2007). The progeny of the asymmetric divisions are a smaller self-renewing meristemoid cell and a larger stomatal lineage ground cell (SLGC)(Fig.1). The meristemoid will divide asymmetrically a few times before differentiating into a guard mother cell (GMC)(Fig.1). The transition from meristemoid to GMC is controlled by the bHLH MUTE, which is closely related to SPCH. mute mutant epidermis is also without stomata, but asymmetrically divided stomatal lineage cells are present, including aborted meristemoids (MacAlister et al. 2007; Pillitteri et al. 2008; Pillitteri and Torii 2007). The GMC will then divide symmetrically one time to form the paired guard cells of a stoma (Fig. 1). This final step is controlled by a third bHLH, FAMA, which is closely related to SPCH and MUTE (Ohashi-Ito and Bergmann 2006). In fama mutants stomatal lineage cells fail to progress past GMC state, though, rather than arresting, the GMC aberrantly divides several times to form "caterpillar" shaped stacks of cells (Ohashi-Ito and Bergmann 2006).
SPCH, MUTE, and FAMA are expressed at specific, non-overlapping times during stomatal development, and their actions are coordinated by a second group of bHLH proteins, SCREAM/ICE1 (SCRM) and SCRM2, that are expressed throughout the stomatal lineage and heterodimerize with SPCH, MUTE, and FAMA (Fig. 1)(Kanaoka et al. 2008). SCRM/SCRM2 proteins appear to function in a dosage-dependent manner. A gain-of-function allele of SCRM, scrm-D, drives constitutive stomatal differentiation in the epidermis, and the effect of the semi-dominant scrm-D mutation is less severe in scrm-D/+ (Kanaoka et al. 2008). Likewise, successive loss of SCRM and SCRM2 results in a range of phenotypes: scrm scrm2 double mutants recapitulate the spch phenotype, loss of scrm with partial loss of scrm2 recapitulates the mute phenotype, and loss of scrm alone resembles the fama phenotype (Kanaoka et al. 2008).
During the development of leaves and other photosynthetic organs, additional stomata are formed in such a way that they will be evenly distributed. It has become clear that a cell-cell signaling mechanism exists and impinges on the transcriptional cascade leading to stomatal cell-type differentiation (Lampard et al. 2008). This review will examine the signaling components that control stomatal spacing in Arabidopsis, with a focus on several related small secretory putative ligands. We will also discuss how the system controlling stomatal patterning is likely fundamental to all land plants.
 Molecular basis of the one-cell spacing rule in Arabidopsis Molecular basis of the one-cell spacing rule in Arabidopsis
One of the first stomatal patterning mutants isolated was too many mouths (tmm), which forms clusters of stomata on leaves, thus violating the one-cell spacing rule (Yang and Sack 1995). The corresponding wild-type TMM protein is expressed solely in the stomatal lineage and encodes a receptor-like protein that is specific to the stomatal lineage, but which likely cannot transmit any signals since it lacks any intracellular domain (Geisler et al. 2000; Nadeau and Sack 2002). Intriguingly, tmm mutants entirely lack stomata on stems, which indicates that TMM regulates stomatal patterning disparately in different organs of the plant: TMM enforces stomatal patterns in leaves but is required for stomatal differentiation in stems (Bhave et al. 2009). Next, the discovery that stomatal clustering results when the more widely expressed ERECTA-family receptor-like kinase (RLK) genes, ERECTA, ERECTA-LIKE 1 (ERL1), and ERECTA-LIKE 2 (ERL2), are mutated, brought potential partners to TMM, providing a means of transmitting a signal into the cell (Shpak et al. 2005). The genetic interactions between TMM and the ERECTA family, however, are complex, and the interaction of TMM with a given ERECTA-family RLK may have an inhibitory effect on signal transduction, rather than promoting such a signal, or TMM may compete for common ligands with ERECTA-family RLKs (Shpak et al. 2005).
Recently, several putative ligands that regulate stomatal spacing have been isolated. All belong to the EPIDERMAL PATTERNING FACTOR-LIKE family (EPFL) of small secretory peptides (Hara et al. 2009), which may be identified by an N-terminal secretory signal sequence and C-terminal end containing six or eight cysteines that likely act in forming intramolecular disulfide bonds (Kondo et al. 2010). It is predicted that the N-terminal end is cleaved to form a mature secreted peptide (Fig. 2)(Hara et al. 2007). Indeed, in the case of one member of the EPFL family, STOMAGEN/EPFL9, the N-terminal end of the mature peptide has been confirmed at the predicted site (Fig. 2a)(Kondo et al. 2010; Sugano et al. 2010). In addition, the exact sites of intramolecular disulfide bonds of STOMAGEN/EPFL9 were also resolved (Fig. 2c)(Kondo et al. 2010). In plants, small cysteine-rich secretory peptides act in cell-cell recognitions, including self-incompatibility response in Brassica and pollen-tube guidance in Trenia (Okuda et al. 2009; Schopfer et al. 1999; Takayama et al. 2000). Although EPFL-family proteins do not share any primary sequence similarity with other cysteine-rich peptidic ligands, the positions and numbers of cysteine patches are somewhat conserved. These make EPFL-family proteins attractive and most likely ligand candidates for TMM and ERECTA-family RLKs.
The first EPFL family member characterized was EPF1, which is normally expressed in late meristemoids, GMCs, and young guard cells (Figs. 1 and 3). EPF1 was found to negatively regulate stomatal density. In strong overexpression of EPF1, the epidermis produces no stomata, but SLGCs that do not accompany a stoma are present. This phenotype is reminiscent of the erecta erl2 double mutant, which has an increase over wild type in the number of SLGCs, although stomata are also formed (Shpak et al. 2005). The similar phenotypes of EPF1 overexpression and erecta erl2 suggest a connection between the ligand EPF1 and the putative receptor ERL1. Loss of function in EPF1 results in an increase in stomatal clustering, so these mutants also violate the one-cell-spacing rule (Hara et al. 2007). Functional TMM and ERECTA-family RLKs as well as the Mitogen-Activated Protein Kinase Kinase Kinase (MAPKKK) YODA are required for the overexpression phenotype of EPF1, suggesting that EPF1 operates upstream in the same pathway as these genes, potentially as a ligand for TMM and ERECTA-family RLKs, which likely transmit a signal through YODA (Bergmann et al. 2004; Hara et al. 2007).
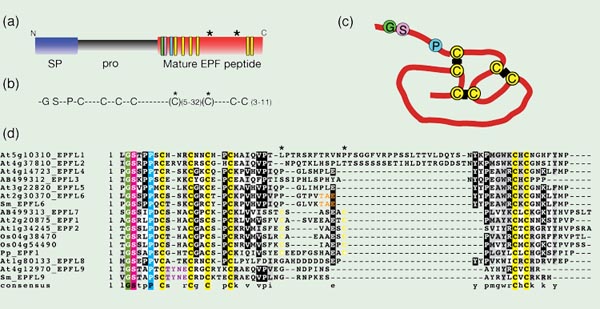 Fig. 2. Epidermal Patterning Factor-Like (EPFL) family of proteins
Fig. 2. Epidermal Patterning Factor-Like (EPFL) family of proteins
(a) Prepropeptide structure, which includes an N-terminal secretory signal sequnece (SP; blue) and propeptide (Pro; gray) that is cleaved from the C-terminal end, which contains the mature EPF peptide (red). The positions of conserved cysteines (yellow), glycine (green), serine (pink), and proline (cyan) residues are highlighted as rectangles. Two asterisks indicate the position of additional cysteine residues found in EPF1, EPF2, and EPFL7.
(b) A consensus sequence among Arabidopsis EPFL-family proteins, which contain conserved glycine (G), serine (S), and proline (P) residues followed by 6 or 8 conserved cysteine residues (C).
(c) Structure of intramolecular disulfide bonds between the six cysteines in STOMAGEN/EPFL9. Modified from Kondo et al., 2010.
(d) Amino acid alignment of all ten EPFL family members in Arabidopsis thaliana (At) and related homologous sequences from the moss Physcomitrella patens (Pp), the lycophyte Selaginella moellendorffii (Sm), and rice, Oryza sativa (Os). EPF1, EPF2, and EPFL7 family members from Arabidopsis, O. sativa, and P. patens are characterized by two extra conserved cysteine residues in yellow (positions marked by asterisks). STOMAGEN/EPFL9 sequences from Arabidopsis and S. moellendorffii are characterized by a TYNE motif in purple, and CHALLAH/EPFL6 sequences from Arabidopsis and S. moellendorffii are identified by a TAE motif in orange.
EPF2 was recently characterized as a small secreted peptide that also negatively regulates stomatal density, though EPF2 acts earlier in stomatal development than EPF1, as reflected by its expression in MMCs and early meristemoids (Figs. 1 and 3)(Hara et al. 2009; Hunt and Gray 2009). The overexpression phenotype of EPF2 is an epidermis entirely devoid of stomata or SLGCs, while epf2 loss of function results in an increase in stomatal cell density and also in non-stomatal epidermal cell density (likely SLGCs), which suggests that EPF2 functions to restrict entry into the stomatal lineage by MMC. Like EPF1, the overexpression phenotype of EPF2 is dependent on functional TMM and ERECTA-family, and pentuple mutant analysis revealed that both EPF1 and EPF2 function upstream of ERECTA, ERL1, and ERL2 (Hara et al. 2009). Because the stomatal clustering phenotype of the epf1 and epf2 mutants is additive in the double mutant, it appears that, while their function is similar during stomatal development, it is nonetheless distinct (Hara et al. 2009; Hunt and Gray 2009). This was further confirmed by promoter-swapping analysis of EPF1 and EPF2 (Hara et al. 2009).
A third EPFL family member, CHALLAH (CHAL), or EPFL6, was found in a sensitized genetic screen, and its loss-of-function mutation rescues the previously mentioned stomataless phenotype of the hypocotyl and inflorescence stem in tmm. Unlike EPF1 and EPF2, CHAL is expressed in a region-specific manner and is not confined to the stomatal lineage or even the epidermis; it is expressed in cells surrounding the vascular elements of the stem (Fig. 3). Similar to EPF1 and EPF2, CHAL acts repressively in stomatal formation, as constitutive strong overexpression results in leaves without stomata (Abrash and Bergmann 2010). However, functional TMM is required for the overexpression phenotype of EPF1 and EPF2, while the overexpression phenotype of CHAL is much more exaggerated in the tmm mutant. When CHAL is overexpressed in tmm and the function of any two ERECTA family members is lost, the overexpression severity is dampened, such that these triple mutants with CHAL overexpression may form stomata. These data indicate that any of the three ER family members may serve as a potential receptor for CHAL and this receptor-ligand interaction is repressed by TMM. The chal loss-of-function mutant phenotype is only revealed in combination with tmm, and chal tmm er triple mutants fail to form stomata on the hypocotyl, reversing the rescue of the chal tmm double combination. chal tmm erl2 triple mutants, on the other hand, produced more hypocotyl stomata than the chal tmm double mutant. The quadruple loss-of-function phentoypes are harder to interpret; because er is epistatic to chal tmm, ERECTA may both transmit a CHAL signal and also protect ERL1 and ERL2 from excess CHAL (Abrash and Bergmann 2010). Therefore, in stems, it is quite likely that EPF1 and EPF2 share the same receptors with CHAL, and that TMM interaction with ERECTA-family RLKs in the leaf and stem may be opposite. Also in the stem, the negative interaction of TMM with the ERECTA-family RLKs may be CHAL dependent, thus potentially explaining the surprisingly opposite tmm phenotypes in the leaf and stem.
The first positive regulator of stomatal density has recently been characterized. Named STOMAGEN, it is EPFL9, another EPFL family member. Like CHALLAH, STOMAGEN is not expressed in the epidermis or in stomatal lineage cells. Instead STOMAGEN is produced in the mesophyll tissues (Fig. 3)(Kondo et al. 2010; Sugano et al. 2010). Opposite to what has been described for EPF1 and EPF2, RNAi knockdown of STOMAGEN results in a nearly stomataless phenotype and overexpression yields stomatal clusters, indicating that STOMAGEN is required to promote stomatal development. However, similar to EPF1 and EPF2, STOMAGEN activity is dependent on SPCH and TMM, since overexpression of EPF1, EPF2, and STOMAGEN in tmm and spch fail to alter their phenotypes. (Hara et al. 2009; Kondo et al. 2010; Sugano et al. 2010). It is possible that STOMAGEN antagonizes the negative actions of EPF1 and EPF2, since application of excess STOMAGEN fails to further increase stomatal density in the epf1 epf2 double mutant (Kondo et al. 2010). This suggests the exciting possibility that STOMAGEN, EPF1, and EPF2 directly compete for the same receptors. On the other hand, RNAi silencing of STOMAGEN reverses the increase in stomatal density in epf1, in epf2, and in epf1 epf2, which indicates that STOMAGEN acts independently of EPF1 and EPF2. The increase in non-stomatal-cell density of the epf2 mutant is not reversed in the STOMAGEN RNAi knockdown lines, which indicates that entry into the stomatal lineage by the MMC is negatively regulated by EPF2 and that STOMAGEN is dependent on EPF2 in regulating the formation of SLGCs. However, STOMAGEN's influence over promoting differentiation of guard cells once they have entered the stomatal lineage is independent of both EPF1 and EPF2 (Sugano et al. 2010).
Because STOMAGEN and CHALLAH are not expressed in the epidermis but instead in the internal tissues (Abrash and Bergmann 2010; Kondo et al. 2010; Sugano et al. 2010), their discoveries reveal the presence of non-cell-autonomous cell-cell communication mediated by EPFL-family signaling ligands not only within the epidermal layer but also across tissue layers, thus expanding our view of the control of stomatal patterning into three dimensions. The inter-cell-layer control of stomatal patterning may ensure the coordinated differentiation of the photosynthetic tissues, such as leaf mesophyll and stem cortex, where carbon-fixation actively occurs, with stomatal numbers and distribution necessary for gas exchange.
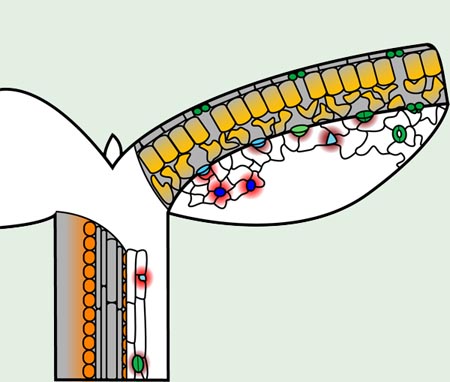
Fig. 3. Localization of EPFL family peptides active in stomatal patterning
In the epidermis, EPF2 (red) is produced by MMCs (blue), while EPF1 (dark red) is produced by meristemoids (cyan), guard mother cells (light green), and young guard cells (green). Mature guard cells do not produce either EPF peptide. Both EPFs suppress stomatal lineage identity in neighboring cells. CHALLAH (orange), which also restricts stomatal differentiation, originates only in cells surrounding the vascular bundle in stems and appears to diffuse to the epidermis. STOMAGEN (amber) is produced in the mesophyll and diffuses to the epidermis to promote stomatal development. Gray areas delineate cross-sections showing interior cells.
In summary, of the 10 to 11 EPFL family members identified in Arabidopsis to date (Abrash and Bergmann 2010; Hara et al. 2009), four have been studied in detail. Two (EPF1 and EPF2) are expressed specifically in the stomatal lineage (one early and one late), and negatively regulate stomatal density. One (CHAL or EPFL6) is subepidermally expressed, organ specific, and also a negative regulator. EPF1, EPF2, and CHAL are likely putative ligands for ERECTA-family RLKs. The fourth (STOMAGEN or EPFL9) acts non-cell-autonomously to promote stomatal development. What is the receptor for STOMAGEN and other EPFL proteins? A compelling case may be made that EPF1 and EPF2 directly compete with STOMAGEN for the same receptors, potentially TMM. Indeed, for all the EPFL family members known to regulate stomatal density, TMM is either required or affects their activity.
 EPFL gene family evolution and future perspectives EPFL gene family evolution and future perspectives
The EPFL gene family is a plant-specific family of secreted peptides, but they are not unique among small peptides. EPFLs are structurally related to defensins, a very large gene family of small cysteine-rich secretory peptides found in animals and plants, which, depending on the group, also have conserved 6 or 8 cysteines that form intramolecular disulfide bonds and function in innate immune response (Boman 1995; Broekaert et al. 1995).
In plants, gene families of small cysteine-rich peptides are highly expanded and diverged (Silverstein et al., 2007), and as mentioned earlier some family members of cysteine-rich peptides act as ligands for cell-cell recognition events during pollination and fertilization (Okuda et al. 2009; Schopfer et al. 1999; Takayama et al. 2000).
A BLAST survey of the genomes of the moss Physcomitrella patens, the lycophyte Selaginella moellendorffii, and rice, Oryza sativa, reveals the presence of several members of the EPFL family (a subset of which are presented in Fig. 2d). Members of the EPF1, EPF2, and EPFL7 group are characterized by a total of 8 conserved cysteine residues, including an additional two cysteine residues compared to the rest of the family members. Two such genes were found in rice, and one in Physcomitrella. STOMAGEN/EPFL9 and its homologs in other plant species share a unique TYNE motif, and one gene with this motif was found in Selaginella (Sugano et al. 2010). CHALLAH/EPFL6 has fewer residues that are unique among all the EPFL family members, but one sequence in Selaginella possesses a TAE motif that matches only CHALLAH among all the EPFL members (Fig. 2d). Presence of several EPFL family members in diverse lineages of land plants implies an origin for the EPFL family in the ancestor to all land plants.
Land plants, including Physcomitrella patens, have conserved master regulatory bHLH proteins that likely direct stomatal cell fate. In addition, homologs for TMM and YODA also exist in P. patens, which indicates that the genes required for both stomatal spacing mechanisms and cell-state specification originated in the land plant ancestor (Peterson et al. 2010). Now, with the finding that basal plant species possess homologs of EPF1, EPF2, and STOMAGEN, we can begin to speculate that the control of stomatal density, having both positive and negative inputs via secreted peptide ligands, is part of the original mechanism of stomatal patterning.
>The regulation of stomatal patterning via small secreted ligands in Arabidopsis is emerging as a complex story of cell-cell signaling that incorporates aspects of inter-cell-layer communication, regional specificity, and potential competitive interaction between positive and negative signaling ligands on the same receptor complex. It will be interesting to discover the function of the other 8 members of the Arabidopsis EPFL family, and to see whether any other members participate in aspects of stomatal patterning. At least EPFL4 and EPFL5 have the potential to suppress stomatal differentiation, since their ectopic overexpression led to decreased numbers of stomata (Hara et al. 2009). Careful loss-of-function studies and higher-order mutant analysis might reveal their precise and potential role in stomatal development. Likewise, in other plants with variant stomatal patterning mechanisms (Peterson et al. 2010), how are EPFL family members participating in diverse modes of stomatal patterning? Comparative studies of stomatal patterning coupled with functional analysis of EPFL-family genes might unravel the underlying molecular basis of the evolution and diversity of stomatal patterning.
References
Abrash EB, Bergmann DC (2010) Development 137:447-455
Bergmann DC, Lukowitz W, Somerville CR (2004) Science 304:1494-1497
Bhave NS, Veley KM, Nadeau JA, Lucas JR, Bhave SL, Sack FD (2009) Planta 229:357-367
Boman HG (1995) Annu Rev Immunol 13:61-92
Broekaert WF, Terras FR, Cammue BP, Osborn RW (1995) Plant Physiol 108:1353-1358
Casson, S, Gray, JE (2008) New Phytol 178:9-23
Geisler M, Nadeau J, Sack FD (2000) Plant Cell 12:2075-2086
Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T (2007) Genes Dev 21:1720-1725
Hara K, Yokoo T, Kajita R, Onishi T, Yahata S, Peterson KM, Torii KU, Kakimoto T (2009) Plant Cell Physiol 50:1019-1031
Hunt L, Gray JE (2009) Curr Biol 19:864-869
Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu JK, Torii KU (2008) Plant Cell 20:1775-1785
Kondo T, Kajita R, Miyazaki A, Hokoyama M, Nakamura-Miura T, Mizuno S, Masuda Y, Irie K, Tanaka Y, Takada S, Kakimoto T, Sakagami Y (2010) Plant Cell Physiol 51:1-8
Lampard GR, Macalister CA, Bergmann DC (2008) Science 322:1113-1116
MacAlister CA, Ohashi-Ito K, Bergmann DC (2007) Nature 445:537-540
Nadeau JA, Sack FD (2002) Science 296:1697-1700
Ohashi-Ito K, Bergmann DC (2006) Plant Cell 18:2493-2505
Okuda S, Tsutsui H, Shiina K, Sprunck S, Takeuchi H, Yui R, Kasahara RD, Hamamura Y, Mizukami A, Susaki D, Kawano N, Sakakibara T, Namiki S, Itoh K, Otsuka K, Matsuzaki M, Nozaki H, Kuroiwa T, Nakano A, Kanaoka MM, Dresselhaus T, Sasaki N, Higashiyama T (2009) Nature 458:357-361
Peterson KM, Rychel AL, Torii KU (2010) Plant Cell 22: 296-306
Pillitteri LJ, Bogenschutz NL, Torii KU (2008) Plant Cell Physiol 49:934-943
Pillitteri, LJ, Sloan, DB, Bogenschutz, NL, Torii, KU (2007) Nature 445:501-505
Pillitteri LJ, Torii KU (2007) Bioessays 29:861-870
Raven JA (2002) New Phytologist 153:371-386
Sachs T (1991) Pattern formation in plant tissues. Cambridge University Press, Cambridge, UK
Schopfer CR, Nasrallah ME, Nasrallah JB (1999) Science 286:1697-1700
Shpak ED, McAbee JM, Pillitteri LJ, Torii KU (2005) Science 309:290-293
Silverstein, KA, Moskal, WA, Jr, Wu HC, Underwood, BA, Graham, MA, Town, CD, VandenBosch, KA (2007) Plant J 51;262-280
Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Hara-Nishimura I (2010) Nature 463:241-244
Takayama S, Shiba H, Iwano M, Shimosato H, Che FS, Kai N, Watanabe M, Suzuki G, Hinata K, Isogai A (2000) Proc Natl Acad Sci U S A 97:1920-1925
Yang, M, Sack, FD (1995) Plant Cell 7:2227-2239
**This is the original, autor-edited version of "Rychel, A.L., Peterson, K.M., and Torii, K.U. (2010) Plant Twitter: Ligands under 140 amino acids enforcing stomatal patterning" published in Journal of Plant Research (copy right, Springer)***
|