RESEARCH |
Single molecule techniques have uncovered the working principles of classic motor proteins, such as myosin, kinesin, and ATP synthase, which are responsible for muscle contraction, intracellular transport, and energy metabolism, respectively. However, these powerful methods have only scarcely been applied to study the mitotic spindle machinery. We are developing in vitro, cell-free motility assays using purified spindle components and bringing advanced single molecule techniques into this research area. Much of our work is carried out in collaboration with other members of the Seattle Mitosis Club, especially the laboratories of Sue Biggins, Trisha Davis, and Linda Wordeman. We focus on many areas fundamental to spindle function: |
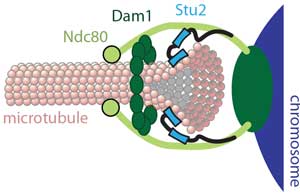
How are chromosomes coupled to the tips of spindle microtubules? Chromosomes attach via kinetochores to the tips of spindle microtubules, sustaining tensile load continuously, even as the tips assemble and disassemble under their grip. Uncovering how such strong yet dynamic tip-coupling occurs is crucial for understanding mitosis. We found that the Ndc80 complex, a primary kinetochore component conserved across eukaryotes, is sufficient by itself to form such attachments (pdf). Individual Ndc80 complexes form 'slippery' (i.e., diffusive) bonds with the microtubule, supporting a biased diffusion mechanism where ensembles of Ndc80 permit thermally-driven and externally-forced movement along the filament, while resisting complete detachment from the tip. The strength of Ndc80-based coupling is enhanced by addition of the Dam1 complex (pdf) and Stu2 (pdf) in yeast, or by the Ska complex (pdf) in humans. Enhancement by Dam1 requires oligomerization (pdf), likely into a microtubule-encircling ring. According to the popular 'conformational wave' model, protofilaments might pull on the Dam1 ring as they curl outward from a disassembling microtubule tip (pdf). We showed that curling protofilaments carry a large amount of energy as bending strain (pdf), and that their work output can be tuned biochemically and vary with species (pdf). Recently, we reconstituted tip attachments using seven recombinant yeast kinetochore subcomplexes that span all the way from centromeric nucleosomes to dynamic microtubule tips (pdf). These observations provide a foundation for studying how kinetochores drive chromosome movement, how they sense and correct attachment errors, and how they regulate the cell cycle. |
 |
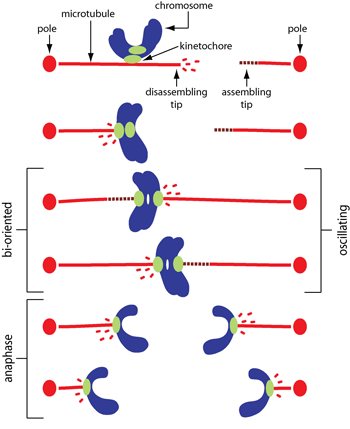 |
How are chromosome movements controlled? Microtubules are inherently stochastic, constantly growing or shortening and switching randomly between these two states – yet they drive exquisitely ordered movements during mitosis. To do so, the filaments must be regulated in response to tensile force. Tension-dependent regulation was assumed to occur via indirect mechanisms, where forces at the kinetochore-microtubule interface modulate the activity of microtubule-modifying enzymes. However, we found that the behavior can be reconstituted without complicated signaling pathways or microtubule-modifying enzymes. In collaboration with Trisha Davis' lab, we developed a motility assay using the Dam1 complex, which lacks any motor or enzymatic activity but forms load-bearing attachments to dynamic microtubule tips (pdf). Then we used a feedback-controlled laser trap to show how tension strongly biases the microtubules toward elongation (pdf). Later, in collaboration with Sue Biggins' lab, we reconstituted tip-coupling using native kinetochores and found the same tension-dependent effects (pdf). Recently, we showed that despite their highly variable growth rates, multiple microtubules can be tightly coordinated with each other merely by coupling their tips to a shared mechanical load (pdf). Altogether, our work in this area has uncovered mechanoregulatory principles that are likely to govern cytoskeletal filament dynamics wherever they form tension-bearing tip attachments, such as at kinetochores, spindle poles, focal adhesions and other filament capture sites. |
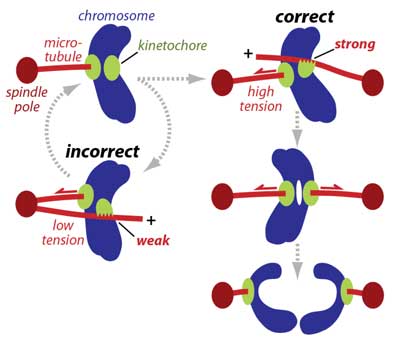
How are aberrant attachments sensed and corrected? During mitosis, accurate segregation requires every pair of replicated sister chromatids to become properly 'bioriented', with their kinetochores attached to the plus ends of microtubules emanating from opposite spindle poles. Improper attachments are unstable and release quickly, giving another chance for proper attachments to form, whereas bioriented attachments are selectively stabilized. It was widely assumed that spindle tension stabilizes bioriented attachments solely by suppressing the destabilizing activity of a kinase called Aurora B. We discovered a more fundamental mechanism: Working with Sue Biggins' lab, we reconstituted kinetochore-microtubule plus-end attachments and showed that tension stabilizes them directly, in the absence of Aurora (pdf). This behavior is analogous to the catch-bonds that enhance cell-cell adhesion, and had not previously been considered for kinetochore-microtubule interactions (pdf). The catch bond-like behavior works in conjunction with the Aurora-based error correction system, which we showed is also regulated directly by tension (pdf). Recently, we discovered that kinetochores grip the sides of microtubules with directionally asymmetric strength (pdf), an effect which probably promotes accuracy during early mitosis, even before both sisters have found plus ends (see diagram at right). Altogether, our work in this area has revealed new mechanisms that ensure the accuracy of chromosome segregation by helping to correct and avoid attachment errors. |
 |
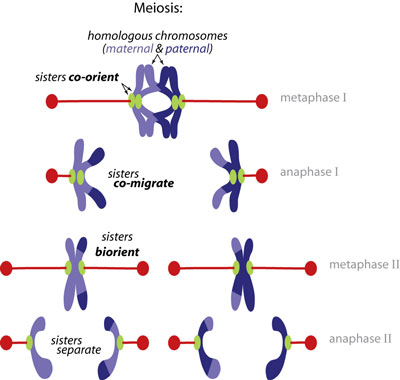 |
How is sister co-migration ensured during meiosis? The cardinal rule of normal mitotic cell division is that replicated sister chromatids must always separate from one another to ensure that complete genomes are distributed to each daughter cell. Sophisticated mechanisms have evolved to impose this rule during mitosis. Strikingly, however, the rule is broken during meiosis, the specialized division that produces eggs and sperm. For accurate meiosis, the sisters must instead co-migrate together during meiosis I, separating from their homologous partners, and only splitting apart later, in meiosis II. Failures in this process are the cause of Down syndrome and other chromosomal birth defects. Studies in yeast suggested that sister kinetochores may be directly fused by a meiosis I-specific factor called monopolin. Working with Adele Marston's lab, we tested this model directly by isolating native meiotic and mitotic yeast kinetochores, reconstituting their function in vitro, and measuring their strength and composition at the single particle level (pdf). Our findings explain why sisters co-orient rather than biorient during meiosis I, which is a conserved behavior underlying Mendelian inheritance. |
New biophysical tools for studying kinetochores, microtubules, and spindle poles. The question of how a single kinesin motor molecule can move unidirectionally along a microtubule without detaching puzzled biologists for fourteen years after the discovery of this property, despite intense study by many groups. I (Chip) showed that it does so by an asymmetric hand-over-hand mechanism (pdf). For my work with kinesin, I co-developed computer-controlled laser traps to record the stepwise, 8-nanometer movements of single kinesin molecules (pdf). Based on these prototypes, my trainees and I in the Asbury lab have built many new instruments (pdf, pdf), and developed new in vitro assays (pdf) specifically for studying how kinetochores harness dynamic microtubules to produce motion and force, and how this viital activity is regulated. Besides kinetochores and microtubules, spindle poles are the other central players in mitosis. We made the first measurements of attachment strength between microtubules and spindle poles (pdf) and more recently showed that pivoting flexibility at the microtubule-pole interface is important for spindle assembly (pdf). |
 |
Home | Research | People | Publications | Photos | Positions | News | Links |
 |
 |