Research Focus
Our lab investigates how cells are influenced by mechanical interactions at the micro and nanoscale. To pursue these goals, we develop new tools (micro/nanodevices, quantitative image analysis, and computational models), which we use to understand the underpinnings of biomechanics and mechanobiology. The greater impact of our work is to delineate how cell mechanics affects cardiovascular disease in order to catalyze new strategies for treatment. By working at the intersection of mechanics and biology, we are also developing an increased understanding of soft, active, and multifunctional materials.
Specifically, we are interested in how biophysical forces, adhesivity, spatial organization, internal structure, and material properties affect the behavior of cells. Since cells are the basic building blocks of organisms, if we can better understand cellular mechanotransduction processes, which are how cells detect and recognize these mechanical cue, then it would be possible to integrate, simulate, and study these interactions at larger scales, e.g. tissue or organ systems.
New Toolsets for Cell Mechanics
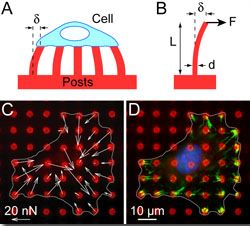 A major challenge in the field of cell mechanics is the lack of tools that are appropriate for studying cells. To address this need, we have focused on the development of new experimental approaches and computational models to study cell mechanics. We have innovated 3D tissue systems [Bielawski (2016), Leonard (2018)], novel fluidic platforms [Ting (2012), Taparia (2017)], magnetic approaches [Sniadecki (2007), Sniadecki (2008), Bielawski (2015), Bielawski (2016)], and nanotechnology [Yang (2007), Ting (2011), Feghhi (2016)] for cell mechanics studies. In addition, we examined cell mechanics using computational models that incorporate fundamental laws and working principles for muscle, mechanics, and biochemistry [Han (2011), Milan (2016), Han (2016)]. These models enable us to postulate on the underlying mechanisms that govern how cells respond to mechanical cues and how they regulate their cellular forces. Lastly, we have made efforts to disseminate our methods and protocols for silicone microposts with a spirit of openness [Sniadecki (2007), Desai (2007), Sniadecki (2014),Beussman (2015)]. As a result, the use of microposts has been adopted by many labs across the world for their own work on cell mechanics.
A major challenge in the field of cell mechanics is the lack of tools that are appropriate for studying cells. To address this need, we have focused on the development of new experimental approaches and computational models to study cell mechanics. We have innovated 3D tissue systems [Bielawski (2016), Leonard (2018)], novel fluidic platforms [Ting (2012), Taparia (2017)], magnetic approaches [Sniadecki (2007), Sniadecki (2008), Bielawski (2015), Bielawski (2016)], and nanotechnology [Yang (2007), Ting (2011), Feghhi (2016)] for cell mechanics studies. In addition, we examined cell mechanics using computational models that incorporate fundamental laws and working principles for muscle, mechanics, and biochemistry [Han (2011), Milan (2016), Han (2016)]. These models enable us to postulate on the underlying mechanisms that govern how cells respond to mechanical cues and how they regulate their cellular forces. Lastly, we have made efforts to disseminate our methods and protocols for silicone microposts with a spirit of openness [Sniadecki (2007), Desai (2007), Sniadecki (2014),Beussman (2015)]. As a result, the use of microposts has been adopted by many labs across the world for their own work on cell mechanics.
Cardiomyocyte Mechanobiology
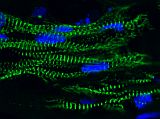 Our work has made impact in understanding the structure and contractile function of cardiomyocytes. Using microposts to measure force and define substrate stiffness, we found that neonatal rat ventricular cardiomyocytes increase their contractile output in response to substrate stiffness [Rodriguez (2011)]. Following on this work, we discovered that cardiomyocytes reduce their myofibril structure and contractile power to counterbalance the effect of reagents that increase actin-myosin force generation, indicating another role for mechanotransduction in cardiac development [Rodriguez (2013)]. Currently, we focus our research to human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CM) [Rodriguez (2014)]. Micropost arrays are a functional assay for hiPSC-CMs with which we can find novel treatments for maturating these cells for heart regeneration and cardiology studies [Yang (2014), Kuppusamy (2015)]. We have also built engineered heart tissues (EHT), which are 3D, tissue-engineered construct comprised of hiPSC-CMs grown in a fibrin gel between a pair of millimeter-scale, silicone posts. We invented a novel, magnetic sensing approach using the silicone posts to measure the contractile activity of EHTs and demonstrated that this approach has potential usefulness in pharmacology studies [Bielawski (2016)]. In addition, we have examined the mechanotransduction response of EHTs to afterload and found that it promotes their maturation, while high levels of afterload drive the onset of pathological markers [Leonard (2018)].
Our work has made impact in understanding the structure and contractile function of cardiomyocytes. Using microposts to measure force and define substrate stiffness, we found that neonatal rat ventricular cardiomyocytes increase their contractile output in response to substrate stiffness [Rodriguez (2011)]. Following on this work, we discovered that cardiomyocytes reduce their myofibril structure and contractile power to counterbalance the effect of reagents that increase actin-myosin force generation, indicating another role for mechanotransduction in cardiac development [Rodriguez (2013)]. Currently, we focus our research to human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CM) [Rodriguez (2014)]. Micropost arrays are a functional assay for hiPSC-CMs with which we can find novel treatments for maturating these cells for heart regeneration and cardiology studies [Yang (2014), Kuppusamy (2015)]. We have also built engineered heart tissues (EHT), which are 3D, tissue-engineered construct comprised of hiPSC-CMs grown in a fibrin gel between a pair of millimeter-scale, silicone posts. We invented a novel, magnetic sensing approach using the silicone posts to measure the contractile activity of EHTs and demonstrated that this approach has potential usefulness in pharmacology studies [Bielawski (2016)]. In addition, we have examined the mechanotransduction response of EHTs to afterload and found that it promotes their maturation, while high levels of afterload drive the onset of pathological markers [Leonard (2018)].
Platelet Biomechanics
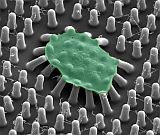 We developed an approach to measure the forces of platelets using micropost arrays, which to the best of our knowledge, was the first method to measure platelet forces at the microscale [Liang (2010)]. We used this approach to find that inhibitors of nonmuscle myosin IIA, Rho kinase, and myosin light chain kinase can inhibit platelet forces [Feghhi (2016)]. We developed a nanoscale version of our post arrays, i.e. nanoposts, to measure the forces of individual platelets and found that a receptor unique to platelets, glycoprotein Ib-IX-V (GPIb-IX-V), can transmit cytoskeletal forces [Feghhi (2016)]. This finding indicates that there are non-integrin receptors that transmit cellular force and can mediate platelet adhesion. Using the micropost technology, Dr. Sniadecki formed a spin-out company, Stasys Medical Corporation to make clinical impact in the detection of platelet dysfunction in cardiology patients and detect bleeding risk in trauma patients.
We developed an approach to measure the forces of platelets using micropost arrays, which to the best of our knowledge, was the first method to measure platelet forces at the microscale [Liang (2010)]. We used this approach to find that inhibitors of nonmuscle myosin IIA, Rho kinase, and myosin light chain kinase can inhibit platelet forces [Feghhi (2016)]. We developed a nanoscale version of our post arrays, i.e. nanoposts, to measure the forces of individual platelets and found that a receptor unique to platelets, glycoprotein Ib-IX-V (GPIb-IX-V), can transmit cytoskeletal forces [Feghhi (2016)]. This finding indicates that there are non-integrin receptors that transmit cellular force and can mediate platelet adhesion. Using the micropost technology, Dr. Sniadecki formed a spin-out company, Stasys Medical Corporation to make clinical impact in the detection of platelet dysfunction in cardiology patients and detect bleeding risk in trauma patients.
Endothelial Mechanotransduction
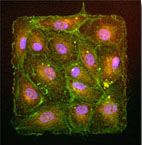 Our research has made key findings on endothelial cells and mechanotransduction at their intercellular junctions. Using mechanical approaches, we helped to find that endothelial proliferation was influenced by regions of high tension within a monolayer of endothelial cells [Nelson (2005)]. We found that the tugging force between pairs of endothelial cells contributed to the size of their adherens junctions [Liu (2010)]. In addition, we found that intercellular tension is critical in how endothelial cells regulate their cell-cell junctions for leukocyte diapedesis [Liu (2010)]. We have identified a "coupled" mechanotransduction relationship between intercellular forces and cell-cell junctions in endothelial cells exposed to fluid shear flow [Ting (2012)]. Specifically, we found that laminar flow induces a rise in intercellular tension between endothelial cells, which in turn leads to reinforcement at their cell-cell junctions. This work with microposts and endothelial cells was featured by The Scientist magazine in an article about tools and techniques for measuring cellular forces.
Our research has made key findings on endothelial cells and mechanotransduction at their intercellular junctions. Using mechanical approaches, we helped to find that endothelial proliferation was influenced by regions of high tension within a monolayer of endothelial cells [Nelson (2005)]. We found that the tugging force between pairs of endothelial cells contributed to the size of their adherens junctions [Liu (2010)]. In addition, we found that intercellular tension is critical in how endothelial cells regulate their cell-cell junctions for leukocyte diapedesis [Liu (2010)]. We have identified a "coupled" mechanotransduction relationship between intercellular forces and cell-cell junctions in endothelial cells exposed to fluid shear flow [Ting (2012)]. Specifically, we found that laminar flow induces a rise in intercellular tension between endothelial cells, which in turn leads to reinforcement at their cell-cell junctions. This work with microposts and endothelial cells was featured by The Scientist magazine in an article about tools and techniques for measuring cellular forces.
Summary
Cells use mechanical factors from the outside and inside to guide their collective function, but there is a lack of appropriate tool-sets with which to study these phenomena. Our micro- and nanofabrication techniques and modeling approaches can allow us to measure these mechanical factors, manipulate the physical interactions, and verify our mathematical predictions. By controlling mechanical interactions at the small scale, we strive to build up a working knowledge of biomechanics that guides new approaches in treatment and prevention of diseases.
|
![[ C E L L B I O M E C H A N I C S L A B ]](pix/banners/www Cell Biomechanics Lab.jpg)
![[ C E L L B I O M E C H A N I C S L A B ]](pix/banners/www Cell Biomechanics Lab.jpg)