Research Projects in the Wendy Thomas Laboratory
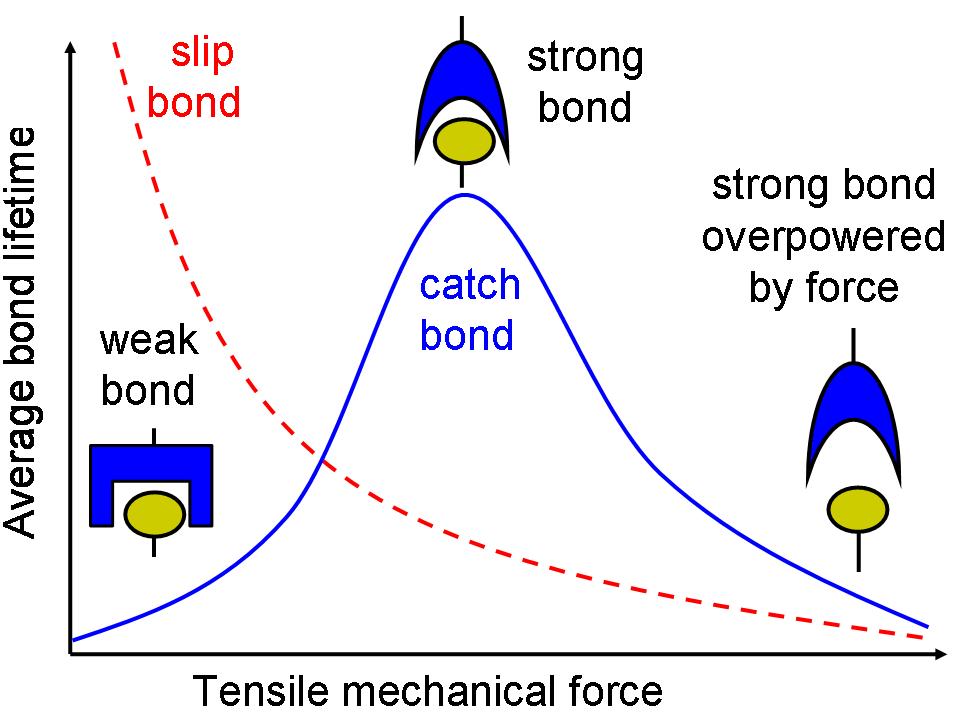
A major project in our lab is learning to engineer catch bonds, which are noncovalent biological adhesive bonds with the counterintuitive property that they are longer-lived when mechanical force is applied to pull them apart. We study a bacterial adhesive protein called FimH that forms catch bonds, in collaboration with Evgeni Sokurenko in the Department of Microbiology at the University of Washington, Viola Vogel at ETH Zurich, Ron Stenkamp at UW Biological Structure, and others. With our collaborators, we are demonstrating that catch bonds can be caused a common means of biochemical regulation called allosteric regulation. By understanding the mechanistic basis of catch bonds, we hope to design therapeutic interventions for infections and diseases that involve catch bonds. We also hope to apply our understanding of catch bond mechanism to design novel catch bonds with unique properties for technological applications. This research is supported by a Bioengineering Research Partnership (BRP) grant from NIH and has funding for a new graduate student.
A second project in our lab involves thrombosis, or blood clots. Most clotting mechanisms do not function well at the high shear stress encountered in arterial wounds. However, one mechanism – binding of platelets to the blood protein called von Willebrand Factor – not only withstands, but actually requires, high shear stress. This mechanism is mediated by binding of the platelet GPIb-alpha domain to the A1 domain of von Willebrand factor. While the structure of this adhesive bond is known from many crystallographic studies (see cartoon on the right), the cause of the shear-activation of this bond is poorly understood. We seek to understand the molecular basis of this shear-activated thrombosis by studying the effects of shear stress and mechanical force on platelet adhesion and aggregation, and on binding and mechanical properties of isolated components. We feel that this work is especially important because the high shear stress needed to induce clotting around wounds is also generated in advanced atherosclerosis, in which plaques restrict blood vessels. The mechanism can thus cause blood clots in restricted vessels, and the clots can then cause a heart attack or stroke. We hope that understanding this mechanism will lead to minimally invasive ways to protect patients with atherosclerosis from deadly blood clots. This project is supported by a Scientist Development Grant from the American Heart Association and may also soon be funded by an R01 grant from the National Heart, Lung and Blood Institute.

We study several bacteria that only bind firmly at high levels of fluid flow, but will roll across the surface at moderate flows and even detach if the flow is low enough. We have studied this behavior mostly in commensal E. coli binding through the adhesive protein FimH, but have also observed it other strains that cause urinary tract infections, endocartitis and diarrhea. One thrust of our research in this area is to understand how the mechanical properties of catch bonds, as well as the long fimbraie that tether the adhesins to the bacteria, are key to the formation and maintenance of biofilms in the presence of flowing bodily fluids. We use genetic and environmental manipulation, force microscopy, and various imaging techniques, and collobarate with microbiologists to understand the role in disease using animal models. We hope to develop methods to prevent biofilms from forming on heart valves and indwelling medical devices such as urinary catheters since these biofilms are resistant to antibiotics.
We have two projects that involve technological applications of regulated adhesion.
One project is what we call our "Actibodies" project, where we design antibody-like recognition proteins that are activatable, meaning that they can be triggered to bind or release their target on cue. We make libraries of these proteins, which can be screended to identify proteins that bind to a target of interest, but in a regulated fashion. Our hypothesis is that certain protein scaffolds are structured so specificity can be arbitrarily changed without affecting the allosteric regulation. We anticipate that the actibody technology could enable improved separation technologies, new capabilities for self-assembled in vitro diagnostic devices, and greater specificity for in vivo molecular imaging or targeted therapeutics. This work is supported by NSF.
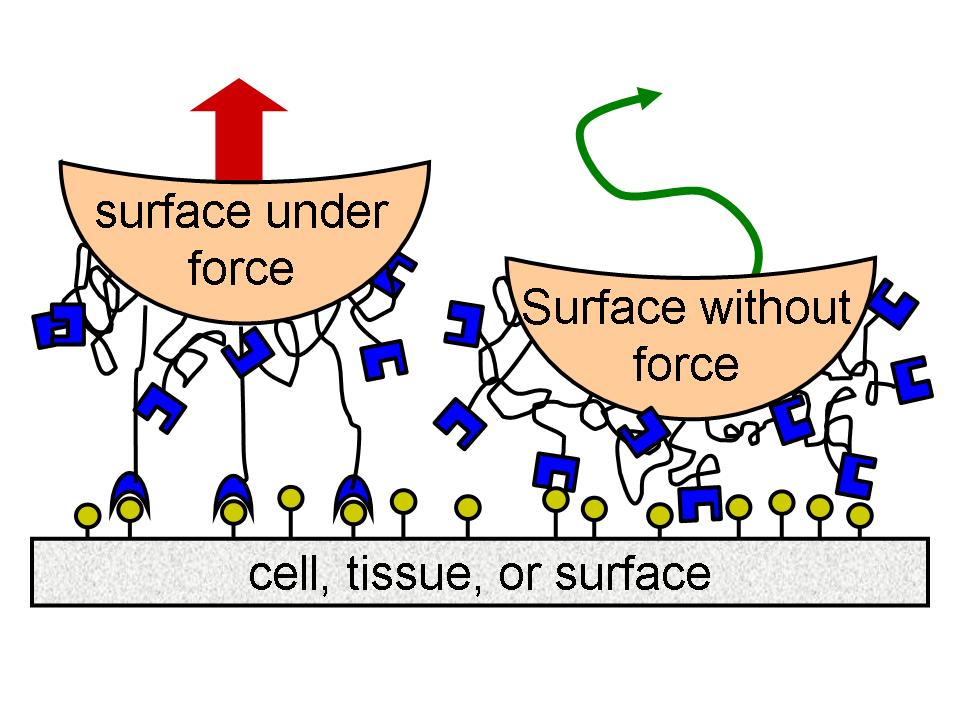
The other project is what we call our "BioCatch" adhesives project. A catch bond is the ultimate in reversible adhesion, gripping strongly under load but detaching when force is removed. We are determining design principles and finding practical applications for mechanically regulated reversible adhesives based on a catch bond used by bacteria that normally live harmlessly in human intestines.Reversible adhesives have many potential technological applications. For example, like geckos walking up a wall, robots need to stick to a surface under load but then let go again. Similarly, robots need to grip and release objects they manipulate. This work is supported by an NSF CAREER award.
.
Many of our research projects involve aspects of protein design since we design mutations and truncations to test out theories about how the proteins we study are regulated. However, our reversible adhesives project involves more extensive protein design, as we redesign proteins to meet new criteria for these technological applications.
We use both Molecular Dynamics Simulations and Rosetta Protein structure prediction techniques to understand both binding specificity and conformational changes in proteins. We make the designed proteins using molecular biology, and test them using various functional assays. We also use directed evolution as a design tool, in which we create libraries of proteins using random mutagenesis and then select clones of interest using high throughput selection techniques.
.
 Wendy Thomas Laboratory
Wendy Thomas Laboratory