|
Research Interests
- Interpretable Machine Learning in Bioinformatics
- Pattern Recognition and Data mining
- Machine Learning and Numerical analysis
- Genome sequencing analysis
- Visualized Medical Data Analysis
Current Research Projects
- iBioML: Collaborative and interpretable machine learning platform for Bioinformatics Research
- Network Motif Analysis
- Evolutionary dynamics through genome sequencing analysis
- VIA-QMI: A software program for immunotherapy research
- Project homepage: Pattern Recognition in Bioinformatics
 Figure 1: iBioML Homepage |
iBioML :Collaborative and interpretable machine learning platform for Bioinformatics Research
iBioML is a collaborative and interactive platform for interpretable machine learning (iML) in Bioinformatics. It promotes the use of iML techniques that are specifically designed for Bioinformatics research, and fosters a community of bioinformatics researchers. |
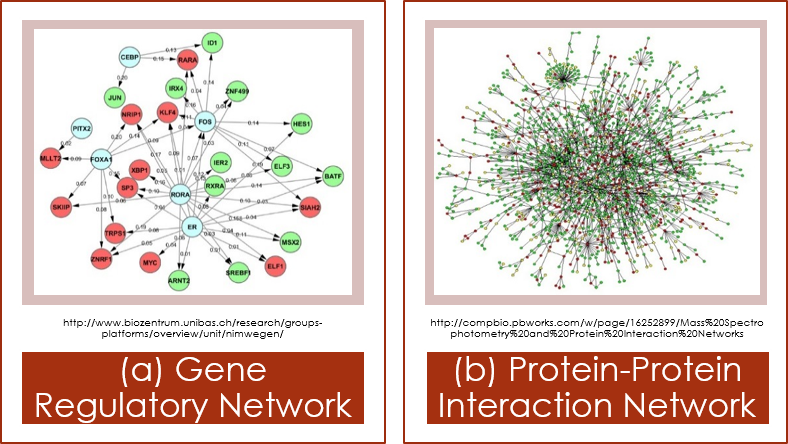 Figure 2: Biological Networks |
Network Motif Analysis
Network motif is a frequent and unique subgraph pattern that can be found in biological networks shown in the left. Network motifs have been applied to find breast-cancer related genes, predict essential proteins, and generate protein interactions. Methods to detect network motifs are categorized into network-based and motif-based approaches. Network-based aproach searches all possible patterns from the input graph, while motif-based approach finds the instances of given query patterns. Fanmod program implements network-based, and Moda implements motif-based approach. |
|
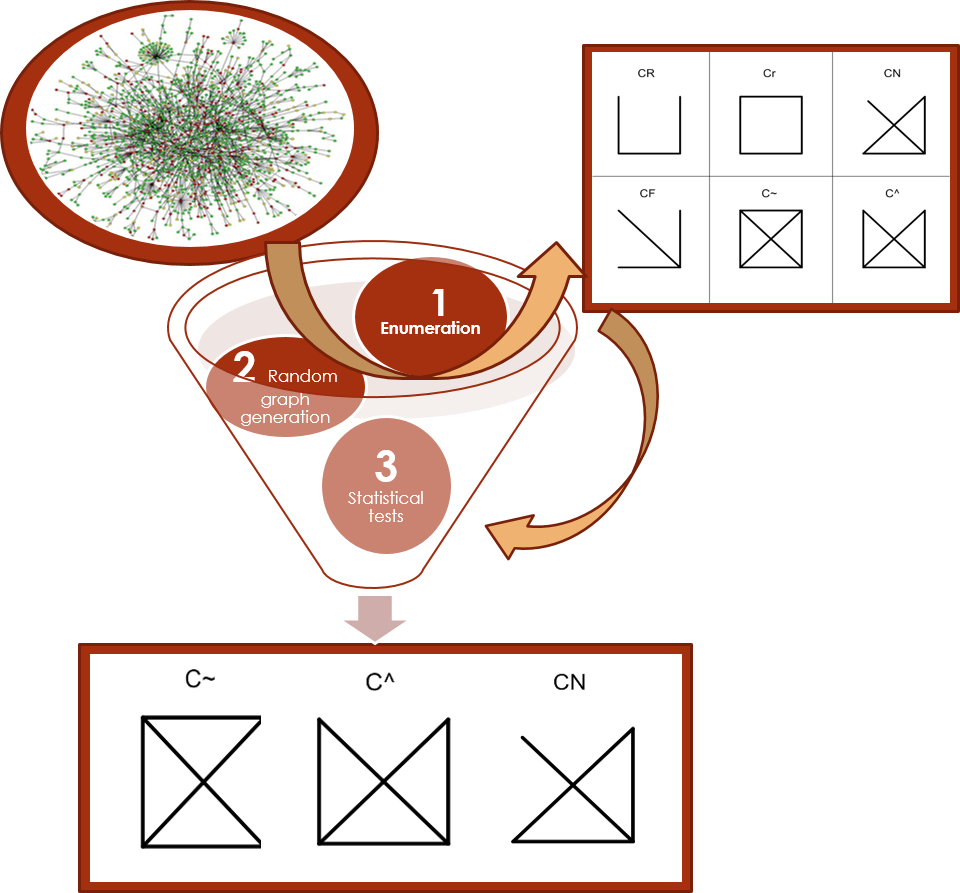
Figure 3: Network Motif Detection |
Examples of network motifs are shown in the left. Out of all non-isomorphic size-4 graph patterns, shown in the upper right, the method can identify frequent and unique patterns (three patterns as in the bottom), and determine those are the network motifs.
Network motif detection process is computationally expensive, and most of programs provide the patterns only without related their instances. We defined a new network motif representation called NemoProfile, which can map pattern to their instances.
We developed network motif libraries (NemoLib) in C++, Java and Pythond, that have NemoProfile functionality. Also we improved the performance of MODA program, which is named as NemoMap. Students have developed web-based network motif detection program using NemoLib.
NemoSuite: Web-based Network Motif Analytic Suite
Download NemoLib: C++, Java, Python
Download NemoMap
Example network file: smallnetwork, bignetwork
|
 Figure 4: Web-based Database System |
Evolutionary dynamics through genome sequencing analysis
We are collaborating with Dr. Kristina Hillesland on Investigating evolution of a microbial mutualism. Sequencing DNA results in many small chunks of DNA strands, which are called reads that combine to make the whole genome. In general, reconstructing the whole DNA sequence from these reads is done in a series of processes including genome alignment, variant calling, and annotation.
We got initial results on list of mutations by the Breseq program. They are available as mutation tables and in a database with muller diagrams (password-protected for now). We are running mutliple tools to detect reliable mutations, and developing an algorithm for detecting structural variants. |
VIA-QMI: A software program for immunotherapy research
We are collaborating with Dr. Stephen Smith on Immonotherapy research. We are developing VIA-QMI (Visualized data analytic tool for QMI), which is a software program that facilitates the Quantitative Multiplex Co-Immunoprecipitation (QMI) platform. The QMI platform generates high-dimentional data that describe the protein-protein interaction network state of the cell. To maximize the usability, and increase data analytic visualization of the process, VIA-QMI is a graphical user interface program that provides a streamlined workflow and an intuitive and interactive interface. |
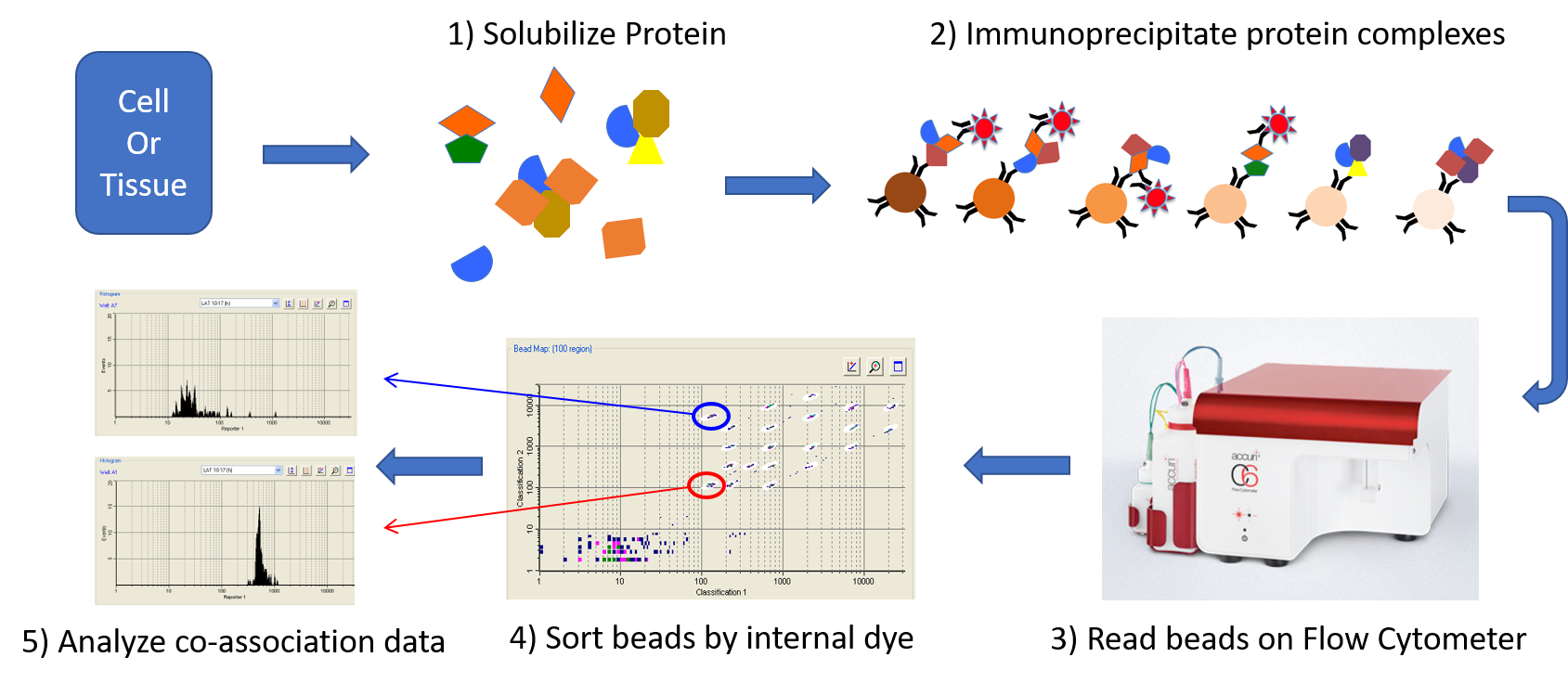
Figure 5: Description of QMI assay |
Award
- R01 CA240985-01A1
Smith (PI), Role: co-investigator
04/15/2020-03/31/2025
Quantitative protein network profiling to improve CAR design and efficacy
National Institutes of Health, $600,156
- Research Assistant Support, Computing and Software Systems Division, University of Washington
- Patterns in a graph to detect structural variants from genome sequencing reads, support for three quarters 2019-2020
- Graphical User Interface Program for Quantitative Multiplex Co-Immunoprecipitation, support for two quarters 2018-2019
- Network Motif in Cloud, support for three quarters 2017-2018
- Reconstruction of Evolutionary Biological Networks, support for two quarters 2016-2017
- CSS Internal Research, Computing and Software Systems Division, University of Washington
- Interpretable Learning in Bioinformatics Applications, Fall 2024 ($8,300)
- Data Morph for Interpretable Deep Learning Applications with Enhanced Computation, Spring 2024 ($8,720)
- Graduate Research Program, Computing and Software Systems Division, University of Washington
- iBioML: Collaborative Infrastructure for Interpretable Machine Learning in Bioinformatics, Winter, Spring, and Summer 2023 ($14,877)
- Interpretable machine learning techniques to identify adaptive
mutations in genomics data, Winter 2022 ($5,000)
- Investigation of genome evolution with network evolution, Spring, and Summer 2020 ($3,751)
- Pattern-based clustering approach to detecting structural variants from next-generation sequencing data, Winter, Spring, and Summer 2019
($5,400)
- Trace mutations in co-evolved organisms with a web-interactive web database system, support for one quarter, Winter 2018 ($3,500)
- Reconstruction of evolutionary PPI networks, Autumn 2016($5,500), Winter 2017 ($5,500), Spring 2017 ($5,500)
-
Machine Learning in Genomics over clouds, Autumn 2014 ($4,400)
-
Motif discovery in MapReduce, Winter 2014 ($2,196)
- PI, Coevolution in-action: Beyond Next Generation Sequencing (NGS) Analysis in Cloud Computing Environments, Microsoft Azure Award, support for one year, 2015, $20,000
- PI, Coevolution in-action: Next Generation Data Sequencing Analysis in Cloud Computing Platforms, Google Cloud Credit, support for one year: Jul 15, 2014 - Jul 14, 2015, $10,000
- NSF RUI 1257525
Hillesland (PI), Role: co-investigator
2013-2016
RUI: Characterization of Coevolution, Adaptation, and Diversification in a Model Microbial Mutualism
National Science Foundation, Division of Environmental Biology, $400,000
Related Links
|



