BACKGROUND:
Many plants flower at a specific time of year to optimize their reproductive success. To predict the timing of seasonal flowering, understanding the governing molecular mechanisms is crucial. Organisms use changes in day length (=photoperiod) and temperature to anticipate upcoming seasonal changes. In my lab, we focus on studying the molecular mechanisms by which plants measure changes in photoperiods. Here I briefly introduce the mechanisms of photoperiodic flowering in Arabidopsis. For further reading regarding this topic, please see the recent reviews [1-4].
Arabidopsis is an ideal system for the study of seasonal flowering mechanisms
![Fig1 external coincid. model.al [Converted]](_Media/fig1-external-coincid-model_med.png)
Many organisms use day length (photoperiod) information to prepare for seasonal changes [5-7]. Photoperiodic response was first described in plants in the early twentieth century [6]. Studies in plants were instrumental in enabling researchers to propose and test conceptual models of day-length measurement. Currently, molecular genetic evidence in plants and animals [8-13] most supports the external coincidence model proposed by Collin Pittendriph in 1964 [14]. In this model, light plays two crucial roles. One is entraining the phase of circadian clock oscillation. The other is activating the clock-controlled key regulator that peaks in late afternoon. Photoperiodic responses are only triggered when the presence of daylight (=the external signal) coincides with peak expression of the key regulator (figure above). Thus, the relationship between presence of light and phase of circadian clock components is important for sensing day-length changes. Desynchronization between daily light periods and internal circadian rhythms causes severe reduction of fitness in cyanobacteria, plants, and animals [15-20].
We have been working on the mechanisms of photoperiodic flowering response in Arabidopsis thaliana, which is currently the most suitable organism for studying molecular mechanisms of day-length measurement. It flowers early under long-day (LD) conditions, while flowering is delayed in short days (SD). This drastic difference in developmental patterns enables us to assess the effects of genetic modification on the photoperiodic response more precisely. Based on our findings as well as others, a detailed model for day-length sensing mechanisms in Arabidopsis has been constituted (see details below).
Flowering pathways in the model plant Arabidopsis
Various environmental factors influence the transition from vegetative to reproductive growth (flowering). The major genetic pathways that control the transition can currently be characterized as photoperiodic, vernalization, autonomous, and gibberellin [10,21,22]. The points of the pathways are interrelated. For instance, in Arabidopsis accessions with a functional FLOWERING LOCUS C (FLC) gene (the key regulator of both the vernalization and autonomous pathways [23,24]), photoperiod sensitivity is suppressed unless plants are vernalized. Without vernalization, FLC MADS-box transcription factor directly suppresses the expression of FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) [25-27]. Eventually, the pathways converge to induce the same floral meristem identity genes, such as LEAFY (LFY), APETALA1 (AP1) and other genes [28,29], and lead to flowering. Ambient temperature changes also affect expression of FT [30,31].
A current mechanistic model for day-length measurement in Arabidopsis
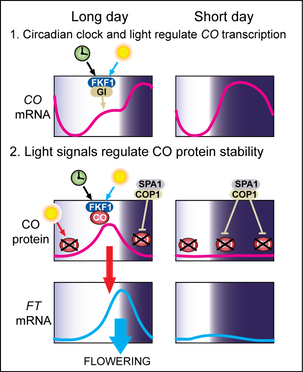
The LD photoperiod promotes flowering in Arabidopsis mainly through the function of CONSTANS (CO) and FT proteins [32-36]. CO is a phloem-specific transcription activator of FT [35,37-39] (see the expression pattern of CO below). Since daytime CO protein expression occurs only when days are long, FT is induced in LD, resulting in earlier flowering [35,36,40] (figure left). To accurately measure differences in day length, circadian clock-dependent timing regulation of CO is a crucial mechanism [35,36]. We demonstrated that the timing of formation of the FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1) and GIGANTEA (GI) protein complex determines the timing of daytime CO gene expression in LD [41]. GI is a large nuclear protein without known functional domain [42], and FKF1 is a LOV domain containing a blue-light photoreceptor that regulates protein degradation [43,44]. The expression of both FKF1 and GI proteins occurs in the afternoon and is regulated by core clock proteins [42-46]. FKF1-GI complex formation occurs only when the FKF1 LOV domain absorbs blue light [41,44]. The role of the FKF1-GI complex is to remove transcriptional repressors of CO, CYCLING DOF FACTORs (CDFs), in the LD afternoon to facilitate expression of daytime CO [47,48].
Light signals perceived by phytochromes and cryptochromes stabilize CO protein only in LD afternoons when daytime CO expression occurs [40] (figure left). We demonstrated that FKF1 physically binds to CO protein in a blue-light dependent manner and stabilizes CO protein in the afternoon in LD [49]. In the dark, even though CO mRNA is highly accumulated, CO protein is actively degraded at night in both LD and SD by the CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) and SUPRESSOR OF PHYA-105 1 (SPA1) E3 ubiquitin ligase complex [40,50-54]. Together with transcriptional control, posttranslational regulation ensures that CO protein exists only in LD afternoons when FT is induced. Thus, circadian-clock regulated CO transcription and light-mediated CO protein stability regulation are the core of the day-length sensing mechanisms.
FT protein is a major part of florigen (a long-sought flowering-inducing substance) and it is synthesized in the leaf vasculature [55-59]. FT protein is then translocated to the shoot apical meristem to initiate expression of floral identity genes, which regulate floral development [38,57,60]. To selectively induce the expression of FT in LD, restricting CO protein expression to the LD afternoon is essential.
The CO/FT module is highly conserved in angiosperms[61,62]. Both CO and FT orthologs exist in long-day and short-day plants. The CO/FT module similarly plays an important role in flowering-time regulation of especially long-day plants, including eudicots and monocots (such as Arabidopsis, poplar, barley and wheat)[61-64]. Thus, Arabidopsis is a suitable model for eudicots and monocots for analyzing the molecular mechanism of photoperiodic flowering. In addition, studying the photoperiodic flowering mechanism is important for understanding a major plant reproduction mechanism, one that is directly applicable to improvement in crop yields.
Fundings:


National Institute of General Medical Sciences
Bibliography:
1. Andrés, F. and Coupland, G. (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13, 627-639
2. Pose, D., et al. (2012) The end of innocence: flowering networks explode in complexity. Curr Opin Plant Biol 15, 45-50
3. Pin, P.A. and Nilsson, O. (2012) The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ 35, 1742-1755
4. Matsoukas, I.G., et al. (2012) Florigenic and antiflorigenic signaling in plants. Plant Cell Physiol 53, 1827-1842
5. Bunning, E. (1969) Common features of photoperiodism in plants and animals. Photochem Photobiol 9, 219-228
6. Salisbury, F.B. (1985) Photoperiodism. Hortic Rev (Am Soc Hortic Sci) 4, 66-105
7. Saunders, D.S. (1997) Insect circadian rhythms and photoperiodism. Invert Neurosci 3, 155-164
8. Yanovsky, M.J. and Kay, S.A. (2003) Living by the calendar: how plants know when to flower. Nat Rev Mol Cell Biol 4, 265-275
9. Hayama, R. and Coupland, G. (2003) Shedding light on the circadian clock and the photoperiodic control of flowering. Curr Opin Plant Biol 6, 13-19
10. Putterill, J., et al. (2004) It's time to flower: the genetic control of flowering time. Bioessays 26, 363-373
11. Imaizumi, T. and Kay, S.A. (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11, 550-558
12. Yoshimura, T. (2010) Neuroendocrine mechanism of seasonal reproduction in birds and mammals. Anim Sci J 81, 403-410
13. Ikegami, K. and Yoshimura, T. (2012) Circadian clocks and the measurement of daylength in seasonal reproduction. Mol Cell Endocrinol 349, 76-81
14. Pittendrigh, C.S. and Minis, D.H. (1964) The entrainment of circadian oscillations by light and their role as photoperiodic clocks. Am Nat, 261-294
15. Ouyang, Y., et al. (1998) Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci U S A 95, 8660-8664
16. DeCoursey, P.J., et al. (2000) A circadian pacemaker in free-living chipmunks: essential for survival? J Comp Physiol A 186, 169-180
17. Beaver, L.M., et al. (2002) Loss of circadian clock function decreases reproductive fitness in males of Drosophila melanogaster. Proc Natl Acad Sci U S A 99, 2134-2139
18. Sharma, V.K. (2003) Adaptive significance of circadian clocks. Chronobiol Int 20, 901-919
19. Woelfle, M.A., et al. (2004) The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol 14, 1481-1486
20. Dodd, A.N., et al. (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630-633
21. Mouradov, A., et al. (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14 Suppl, S111-130
22. Simpson, G.G. and Dean, C. (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296, 285-289
23. Michaels, S.D. and Amasino, R.M. (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949-956
24. Henderson, I.R., et al. (2003) The need for winter in the switch to flowering. Annu Rev Genet 37, 371-392
25. Hepworth, S.R., et al. (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. Embo J 21, 4327-4337
26. Searle, I., et al. (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20, 898-912
27. Helliwell, C.A., et al. (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46, 183-192
28. Ruiz-Garcia, L., et al. (1997) Different roles of flowering-time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9, 1921-1934
29. Blazquez, M.A. and Weigel, D. (2000) Integration of floral inductive signals in Arabidopsis. Nature 404, 889-892
30. Blazquez, M.A., et al. (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet 33, 168-171
31. Balasubramanian, S., et al. (2006) Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet 2, e106
32. Putterill, J., et al. (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847-857
33. Kobayashi, Y., et al. (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960-1962
34. Kardailsky, I., et al. (1999) Activation tagging of the floral inducer FT. Science 286, 1962-1965
35. Suárez-López, P., et al. (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116-1120
36. Yanovsky, M.J. and Kay, S.A. (2001) Signaling networks in the plant circadian system. Curr Opin Plant Biol 4, 429-435
37. Samach, A., et al. (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613-1616
38. Wigge, P.A., et al. (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056-1059
39. Tiwari, S.B., et al. (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol 187, 57-66
40. Valverde, F., et al. (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003-1006
41. Sawa, M., et al. (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261-265
42. Fowler, S., et al. (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18, 4679-4688
43. Nelson, D.C., et al. (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101, 331-340
44. Imaizumi, T., et al. (2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426, 302-306
45. Mizoguchi, T., et al. (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17, 2255-2270
46. David, K.M., et al. (2006) Arabidopsis GIGANTEA protein is post-transcriptionally regulated by light and dark. FEBS Lett 580, 1193-1197
47. Imaizumi, T., et al. (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293-297
48. Fornara, F., et al. (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17, 75-86
49. Song, Y.H., et al. (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336, 1045-1049
50. Hoecker, U. and Quail, P.H. (2001) The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J Biol Chem 276, 38173-38178
51. Ishikawa, M., et al. (2006) The Arabidopsis SPA1 gene is required for circadian clock function and photoperiodic flowering. Plant J 46, 736-746
52. Laubinger, S., et al. (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133, 3213-3222
53. Jang, S., et al. (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27, 1277-1288
54. Liu, L.J., et al. (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20, 292-306
55. Takada, S. and Goto, K. (2003) TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15, 2856-2865
56. An, H., et al. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131, 3615-3626
57. Corbesier, L., et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030-1033
58. Jaeger, K.E. and Wigge, P.A. (2007) FT protein acts as a long-range signal in Arabidopsis. Curr Biol
59. Mathieu, J., et al. (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17, 1055-1060
60. Abe, M., et al. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052-1056
61. Greenup, A., et al. (2009) The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann Bot 103, 1165-1172
62. Song, Y.H., et al. (2010) Similarities in the circadian clock and photoperiodism in plants. Curr Opin Plant Biol 13, 594-603
63. Böhlenius, H., et al. (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040-1043
64. Distelfeld, A., et al. (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12, 178-184