AKILESH Lab at the University of Washington
Copyright 2023 Shreeram Akilesh.
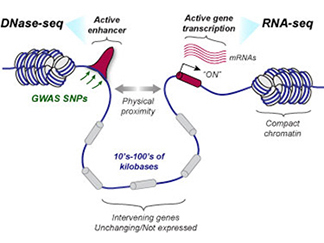
We have generated reference-quality genome-wide functional genomic maps encompassing chromatin-accessibility (DNase-seq), gene expression (RNA-seq) data from primary cultures of human glomeruli, renal cell carcinoma and matched proximal tubule cells. We have also generated and analyzed long-range chromatin conformation data (Hi-C) from human glomeruli and intact human kidney. These first-of-kind epigenomic maps for the kidney have enabled functional annotation of kidney genome-wide association study (GWAS) loci (REF), (REF), (REF), (REF), (REF), (REF). We are leveraging our specialized expertise in kidney anatomy and cellular phenotypes to generate and analyze additional high-resolution datasets from other kidney cell types and anatomic compartments. Our work was highlighted on the cover of the March 2019 issue of JASN and on Science in Seattle. Recently, we have studied the kidney's epigenomic response to injury and corroborated our findings in a 3D "kidney-on-a-chip" system (REF). A recent study of the chromatin architecture and transcriptional differences between kidney cortex and medulla is now in revision. Our kidney functional genomic data is freely available in a UCSC browser instance. Explore our data HERE. Navigate to the "Akilesh-lab-data" tab below the genome browser pane to change configurations or load our previously published datasets, which can be analyzed together!
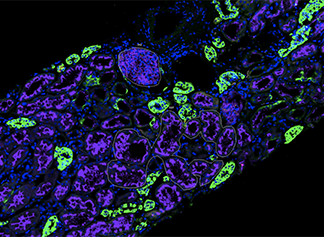
We have had a longstanding interest in developing in situ hybridization technologies (REF), (REF), (REF). Recently, we were the first group to use Digital Spatial Profiling technology on clinically sourced human kidney biopsies to understand mechanisms of virally induced collapsing glomerulopathy (REF). Dr. Akilesh is a Director of the Spatial Biology Core Facility which assists investigators in the design and execution of spatial bioloy experiments. Dr. Akilesh is also a multi-PI on a NIH/NIDDK grant to use this technology to study mechanisms of collapsing glomerulopathy in patients with COVID-19.

In addition to experimental research, our expertise in anatomic and medical renal pathology enables us to view research questions with an eye towards clinical translation (REF), (REF), (REF), (REF), (REF), (REF). We are actively exploring how SARS-CoV-2, the causative agent of COVID-19, affects the kidney using immunohistochemistry, electron microscopy and RNA in situ hybridization (RNA-ISH) (REF), (REF), (REF).
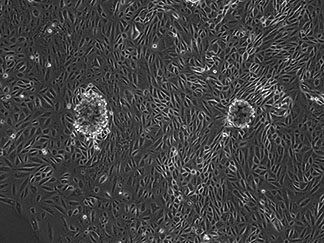
We have extensive experience in the cell biology and genetics of podocytes - the specialized cell of the kidney's filtration unit (the glomerulus). We have linked genetics of familial kidney disease syndromes to the cell biological alterations of podocytes. In collaboration with Dr. Michelle Winn (Duke University), we screened several patients with familial proteinuric kidney disease (focal and segmental glomerulosclerosis, FSGS) for mutations in actin regulatory genes. This lead to the identification of a pedigree with a mutation in the ARHGAP24 gene. We then showed that this gene is developmentally upregulated in mouse podocytes and that it inactivates Rac1 downstream of RhoA activation. Incorporations of patient mutations abrogated this Rac1-inactivation capability (REF). Importantly, this study predicted that if Rac1 was inappropriately activated, podocytes would flatten their cell shape leading to compromise of kidney filter function. We subsequently tested this hypothesis directly by introducing a doxycycline-inducible constitutively active Rac1 transgene into mouse kidney podocytes. Upon induction, Rac1 expressing podocytes flattened their cytoskeleton resulting in proteinuria (REF). Together, these studies have led to a consensus model of podocyte response to injury – in response to various injurious stimuli, podocytes alter their normally RhoA-dominant signaling state to a Rac1-dominant state, resulting in cellular flattening and filter compromise (proteinuria). These studies have been validated and extended upon by several other groups in the field, including in our study of a B-cell mediated model of minimal change disease (REF). We have also recently contributed to a study of transcriptional changes that occur as podocytes undergoing normal aging (REF).
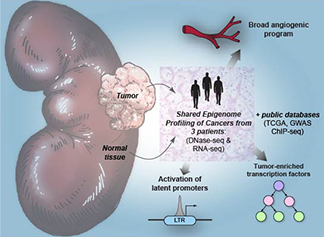
We also study the epigenomics of clear cell renal cell carcinoma (RCC), the most common type of malignant kidney tumor. We recently reported a mechanism for the widespread activation of the potent stem cell transcription factor, OCT4 in RCC (REF). A latent endogenous retroviral long terminal repeat (LTR) element near the OCT4 gene is activated by HIF stabilization that is often seen in RCC. We reported that not only is OCT4 consistently overexpressed, but that it is enriched in open chromatin regions of RCC, thereby directing the transcriptional program of the tumor. Higher OCT4 expression levels in RCC are associated with a poorer prognosis. We are translating these findings using a flexible in vitro mimetic of RCC made with primary tumor cells and endothelial cells (REF). This is the first model of spontaneous angiogenesis towards tumor cells and is amenable to pharmacologic and immunologic interventions. We are funded by the DOD to further advance these studies.
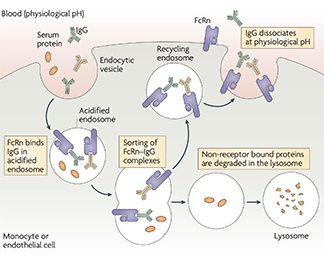
We have contributed significantly to the field of IgG antibody pharmacodynamics and biodistribution through my various studies on FcRn, the neonatal Fc-receptor for IgG. FcRn was initially characterized as the specific receptor that transports maternal IgG antibodies across the placenta to confer passive humoral immunity to the fetus. However, it was also predicted to be expressed in a variety of tissues in the adult where its role was unclear. We contributed to generating the FcRn knockout mouse which demonstrated a markedly reduced serum half-life for IgG antibodies (REF). This proved that FcRn normally rescues IgG antibodies from degradation and is responsible for their long ~21 day half-life in the circulation. An additional prediction from these studies was that FcRn would enhance the serum persistence of pathogenic antibodies and could promote autoimmune disease. We then showed that in a mouse model of antibody-mediated arthritis, FcRn deficiency or inhibition (via high dose, excess IgG, intravenous immunoglobulin – IVIg) reduced disease symptoms and tissue damage (REF). These seminal studies provided a mechanistic rationale for the clinical use of IVIg in the treatment of patients with IgG antibody-mediated autoimmune diseases. Next, we used bone marrow chimeras in mice to demonstrate that bone marrow derived cells (including macrophages) use FcRn to rescue internalized IgG from degradation and return it intact into the circulation (REF). This study and its related review (REF) laid the foundation for understanding the cells and tissue sites responsible for the serum persistence of IgG with immediate practical applications towards the design and production of all monoclonal antibodies and Fc-coupled therapeutics. Our studies of biodistribution had identified that FcRn is expressed in specialized cells (podocytes) of the kidney’s filtration unit. We demonstrated that FcRn knockout mice accumulated IgG at this filtration barrier as they aged, suggesting that FcRn is responsible for preventing the kidney filter from being clogged by common serum molecules (REF). This study was the first to break the established dogma that the kidney filter is completely impervious to large serum molecules; subsequent studies by others have confirmed and extended these findings.
Copyright 2023 Shreeram Akilesh.