Epigenetic and genetic population structure is coupled in a marine invertebrate
Citation
Katherine Silliman and others, Epigenetic and Genetic Population Structure is Coupled in a Marine Invertebrate, Genome Biology and Evolution, Volume 15, Issue 2, February 2023, evad013, https://doi.org/10.1093/gbe/evad013
Abstract
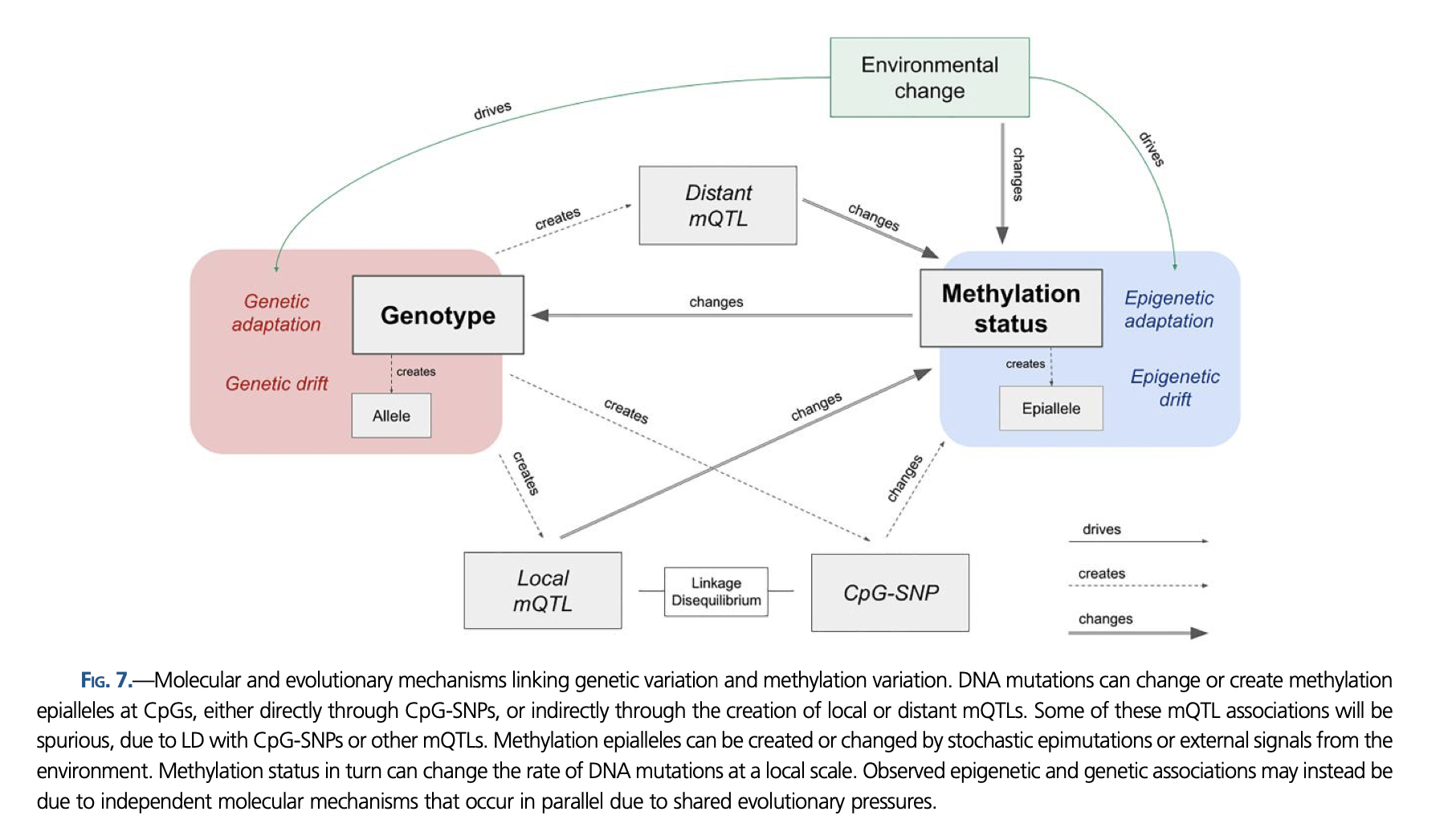
Delineating the relative influence of genotype and the environment on DNA methylation is critical for characterizing the spectrum of organism fitness as driven by adaptation and phenotypic plasticity. In this study, we integrated genomic and DNA methylation data for two distinct Olympia oyster (Ostrea lurida) populations while controlling for within-generation environmental influences. In addition to providing the first characterization of genome-wide DNA methylation patterns in the oyster genus Ostrea, we identified 3,963 differentially methylated loci between populations. Our results show a clear coupling between genetic and epigenetic patterns of variation, with 27% of variation in interindividual methylation differences explained by genotype. Underlying this association are both direct genetic changes in CpGs (CpG-SNPs) and genetic variation with indirect influence on methylation (mQTLs). When comparing measures of genetic and epigenetic population divergence at specific genomic regions this relationship surprisingly breaks down, which has implications for the methods commonly used to study epigenetic and genetic coupling in marine invertebrates.
Significance
We know that genotype and epigenetic patterns are primarily responsible for phenotype, yet there is a lack of understanding to what degree the two are linked. Here, we characterized the degree by which genetic variation and DNA methylation variation are coupled in a marine invertebrate and identified potential mechanisms, with almost a third of the methylation variation attributable to genotype. This study provides a framework for future studies in environmental epigenetics to take genetic variation into account when teasing apart the drivers of phenotypic variation. By identifying methylation variation that cannot be attributed to genotype or environmental changes during development, our results also highlight the need for future research to characterize molecular mechanisms adjacent to genetic adaptation for producing long-term shifts in phenotype.
Data Availability
Code, intermediate analysis files, and genome annotation files used in this study are available in the accompanying repository https://github.com/sr320/paper-oly-mbdbs-gen (https://doi.org/10.5281/zenodo.7083311). The genome assembly can be found at ENA under the accession PRJEB39287 and the raw data are available at NCBI Sequence Read Archive (SRA) at BioProject PRJNA316624. Raw 2b-RAD data are available on SRA at BioProject PRJNA851765. Raw MBD-BS data are available on SRA at BioProject PRJNA849214.