|
Baneyx BioNano Laboratories.
|
|
..Molecular Chaperones
|
|
|
|
|
|
Molecular chaperones are a ubiquitous class of folding modulators that play a central role in the conformational quality control of the proteome by interacting with, stabilizing and remodeling a wide range of nonnative polypeptides. Although constitutively expressed under balanced growth conditions, many chaperones are upregulated upon heat shock or other insults that increase cellular protein misfolding (including heterologous protein expression) and are therefore classified as stress or heat shock proteins (Hsps). Mechanistically, molecular chaperones rely on the differential exposure of structured hydrophobic domains to the solvent to bind nonpolar segments that would normally be buried within the core of their substrates. While there are subtle differences in the composition of client protein recognition sequences (and hence some degree of selectivity in substrate capture), the typical chaperone target is a short unstructured stretch of hydrophobic amino acids flanked by basic residues and lacking acidic residues. The fact that such motifs are common explains why chaperones are so promiscuous.
Molecular chaperones have be divided into three functional subclasses based on their mechanism of action. "Folding" chaperones (e.g., DnaK and GroEL) rely on ATP-driven conformational changes to mediate the net refolding/unfolding of their substrates. "Holding" chaperones (e.g., Hsp33, Hsp31 and IbpB) maintain partially folded proteins on their surface to await availability of "folding" chaperones upon stress abatement. The third class of chaperone (e.g., ClpB) promotes the solubilization of proteins that have become aggregated as a result of stress. A brief description of the main molecular chaperones in the cytoplasm of Escherichia coli is provided below.
|
|
|
|
| .The GroEL-GroES system | back | |
|
|
|
|
|
|
The bacterial chaperonin GroEL and its cochaperone GroES belong to the Hsp60/Hsp10 (aka Cpn60/Cpn10) family of heat shock proteins. Bacterial GroEL is an ≈ 800-kDa hollow cylinder consisting of two seven-subunit rings stacked back to back. GroEL uses a set of hydrophobic residues located at either end of the cylinder to capture non-native substrate and to interact with its co-chaperone GroES, a 70-kDa, dome-shaped homoheptamer. In vivo, binding of client proteins occurs on the ring of GroEL that is not complexed to GroES (known as the trans ring) and involves multiple contacts with the substrate that lead to nanomolar dissociation constants. GroEL can fold proteins up to 60-kDa in size and appears to exhibit a preference for compact intermediates consisting of two or more domains with alpha/beta-folds that are enriched in hydrophobic and basic residues.
GroEL-mediated protein folding is believed to involve the following sequence of events: (1) substrate binding to the nucleotide-free trans ring; (2) binding of 7 ATP molecules and encapsulation by GroES which results in substrate release in an enlarged and now hydrophilic cavity as well as ejection of bound ADP and GroES from the opposite ring; (3) substrate folding timed by the hydrolysis of ATP (≈ 10s); and (4) release of GroES, ADP and either properly folded protein or folding intermediate caused by the binding of 7 ATPs and fresh substrate to the opposite ring. If the ejected substrate still exhibits significant surface hydrophobicity, it will be re-captured by GroEL and the above cycle repeated until a correct conformation is reached.
More information on the GroEL-GroES system is available on Helen Saibil's site.
|
|
 |
|
| ATP-driven confor-mational changes during the GroEL-GroES cycle of action. From Helen Saibil's site |
|
|
| .The DnaK-DnaJ-GrpE system | back | |
|
|
|
|
|
 |
|
|
|
|
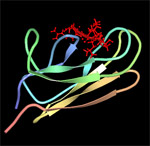
DnaK (Hsp70) is a 69-kDa, monomeric, two-domain protein that plays a central role in de novo protein folding, host protein refolding, translocation and in the general management of the deleterious effects of stress. The DnaK N-terminus domain is responsible for its ATPase activity while the C-terminus domain is involved in substrate binding. The substrate specificity of DnaK has been inferred from the crystal structure of its C-terminal domain and by assessing the ability of the chaperone to bind to immobilized random peptide libraries. DnaK was found to exhibit a preference for heptameric stretches of amino acids composed of a 4-5 residues-long hydrophobic core enriched in leucines and flanked by basic residues. Statistical analyses indicate that these motifs are quite common, occurring every 36 residues on the average protein. It is therefore not surprising that DnaK binds to a large number of structurally and functionally unrelated proteins provided that they are in a nonnative (partially unfolded) state. However, some sites are preferred as binding constants can vary between 5 nM and 5 µM.
|
|
| DnaK-substrate complex. The substrate binding domain of DnaK is shown in complex with peptide NRLLLTG (red) which was selected by phage display. Structure rendered from PDB 1Q5L. |
|
|
|
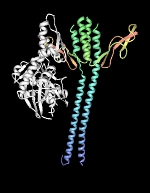
To properly function in vivo, DnaK requires the assistance of two additional cofactors, DnaJ (Hsp40) and GrpE. DnaJ is a 41-kDa protein that triggers ATP-hydrolysis dependent substrate association of partially folded proteins to the substrate binding cavity of DnaK, and contains a conserved J domain that is required for its association with DnaK. DnaJ can independently bind unfolded proteins with low affinity and is believed to scan partially folded substrates in order to direct DnaK to high affinity binding sites. GrpE, a homodimer of 20-kDa subunits, triggers ADP release from DnaK and subsequent substrate release as ATP rebinds DnaK. It is generally accepted that substrate proteins ejected from DnaK either fold into a proper conformation, are recaptured by DnaK-DnaJ for additional cycles of binding and release or are transferred in a partially folded form to GroEL-GroES for subsequent folding.
|
|
|
|
|
 |
|
| DnaK-GrpE complex. The nucleotide exchange factor GrpE is shown in complex with the ATPase domain of DnaK (white). The GrpE dimer is held together by two long alpha-helices from each monomer that terminate into a four-helix bundle involving two helices from each monomer. Structure rendered from PDB 1DKG. |
|
|
| .The Clp disaggregase | back | |
|
|
|
|
|
|
|
|
ClpB belong to the Hsp100 family of ring-forming heat shock proteins whose role is to either degrade unfolded/misfolded proteins, or to solubilize and reactivate aggregated proteins. ClpB, which functions independently of a protease component, is a hexameric ATPase involved in protein disaggregation. This activity is essential for cells to transiently survive extreme thermal stress. Current models hold that the DnaK-DnaJ-GrpE system plays a critical role in the early stages of the disaggregation reaction by facilitating ClpB-mediated extraction of single molecules from aggregates. The captured substrate is next threaded through the 1.6 nm central pore of ClpB where it undergoes net unfolding in a mechanical process that consumes ATP. As it exits the ClpB channel, the unfolded polypeptide may spontaneously refold or be transferred to the DnaK-DnaJ-GrpE and/or GroEL-GroES for chaperone-assisted refolding.
ClpB contains a highly mobile N-terminus domain that surrounds its pore and contributes to enhanced disaggregation activity against certain substrates. The N domain is however dispensable for aggregate solubilization (in fact E. coli synthesizes two versions of ClpB, one containing and one lacking an N domain, from the same mRNA). On the other hand, the ClpB M domain, a coiled coil structure that projects laterally from the top of the ring, appears to play an essential role in the disaggregation process by coupling DnaK-DnaJ-GrpE aggregate "feeding" and ClpB threading motor activity.
|
|
 |
|
| Reconstructed structure of the ClpB hexamer. The N domain is not visible on the reconstruction. The M domain projects outwards from the "top" portion of the hexameric ring. The structure shown is from T. thermophilus and is discussed in more detail by Francis Tsai here. |
|
|
| .Hsp33: a redox-activated holding chaperone | back | |
|
|
|
|
|
|
|
 |
|
|
Hsp33, which was identified on the basis of its thermal induction, is a redox-regulated chaperone that stabilizes proteins unfolded by severe oxidative stress. Under balanced growth conditions, Hsp33 is a reduced monomer that coordinates a zinc atom via four conserved cysteine residues. When cells are exposed to reactive oxygen species – a situation that commonly accompanies heat shock – the cytoplasm becomes more oxidizing and Hsp33 monomers form intramolecular disulfide bonds. Zinc release and conformational rearrangement of the protein's C-terminus into an unstructured form lead to dimerization. The Hsp33 dimer now exhibits chaperone activity and can capture unfolded proteins via its N-terminal substrate binding domains. The thioredoxin and glutaredoxin systems rapidly reduce Hsp33 disulfides in a process that does not cause substrate release but primes the chaperone for fast inactivation. Upon return to nonstress conditions, DnaK-DnaJ engage the bound substrate and refold it alone or with the help of GroEL-GroES.
More information on Hsp33 can be found on Ursula Jakob's site.
|
|
| Redox activation of Hsp33. Activation of Hsp33 holding chaperone function requires monomer to dimer conversion. The picture was modified from that shown in Ursula Jakob's site. |
|
|
| .Hsp31: managing heat and acid stress | back | |
|
|
|
|
|
 |
|
|
|
Hsp31, the hchA gene product, is a heat-inducible homodimeric chaperone belonging to the ThiI/DJ-1/PfpI protein superfamily. Although each Hsp31 monomer contains a poorly accessible catalytic triad responsible for a weak, Zn-dependent aminopeptidase activity, Hsp31 is not a protease. Rather, it functions as a holding chaperone that helps cells handle protein unfolding under extremes of temperature and plays a central role in the ability of starved E. coli to survive acid stress.
The hchA gene is transcribed from dual sigma-D (sigma-70) and sigma-S dependent promoters that are silenced by the nucleoid-binding protein H-NS. Thermal induction of hchA in exponentially growing cells is independent of the alternative heat shock factor sigma-H (sigma-32). Instead, it involves high-level transcription from the sigma-D-dependent promoter upon heat-induced release of H-NS silencing and increased stability of the hchA transcript at high temperatures. Transcription from the sigma-S-dependent promoter dominates when cells enter stationary phase.
The Hsp31 dimer contains a ≈20Å hydrophobic bowl proximal to flexible linker-loop regions that shield large nonpolar patches on either side of the bowl. Temperature-induced motion of the linker-loop domains allows efficient capture of unfolding intermediates by uncovering high affinity binding sites adjacent to the bowl. The linker-loop region may also play a role in substrate ejection by returning to its original position upon stress abatement. Interestingly, although Hsp31 is not an ATPase, its chaperone activity is negatively regulated by ATP binding at high temperature, possibly to coordinate substrate capture with the needs of the chaperone network.
|
|
| Molecular surface of the Hsp31 dimer. A deep acidic canyon connects catalytic triads (arrowhead) in the two monomers. The hydrophobic bowl that serves as a low affinity binding site for partically folded proteins is shown. |
|
|
|
|
 |
|
| Hsp31 mechanism of action. At physiological temperatures, the Hsp31 bowl (cyan) is available for low affinity substrate binding. Heat shock triggers movement of the linker-loop regions (pink/purple) and the exposure of underlying hydrophobic patches (yellow) for efficient substrate capture. |
|
|
| .The IbpA and IbpB small heat shock proteins | back | |
|
|
|
|
|
IbpA and IbpB are two homologous, 16-kDa proteins belonging to the alpha-crystallin (aka small heat shock) protein family of molecular chaperones. They are encoded on a single operon and were first identified as contaminants present in recombinant protein inclusion bodies. IbpB exhibits chaperone function in vitro and forms large amorphous aggregates that dissociate into ≈ 600-kDa oligomers following incubation at high temperatures. IbpA/B are thought to function as holdases that maintain partially folded polypeptides on their surface until stress has abated and the DnaK-DnaJ-GrpE system becomes available for processing of bound substrates. They also intercalate within large protein aggregates that form under stress conditions to facilitate substrate disaggregation and refolding by DnaK-DnaJ-GrpE alone or in collaboration with ClpB.
|
|
|
| | home | research | publications | people | covers | links | teaching | intranet |
Contact: François Baneyx, University of Washington, Department of Chemical Engineering, Box 351750, Seattle, WA Tel: 206-685-7659 Fax: 206-685-3451 E-mail: baneyx@u.washington.edu
© 2008 François Baneyx - All Rights Reserved
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|






