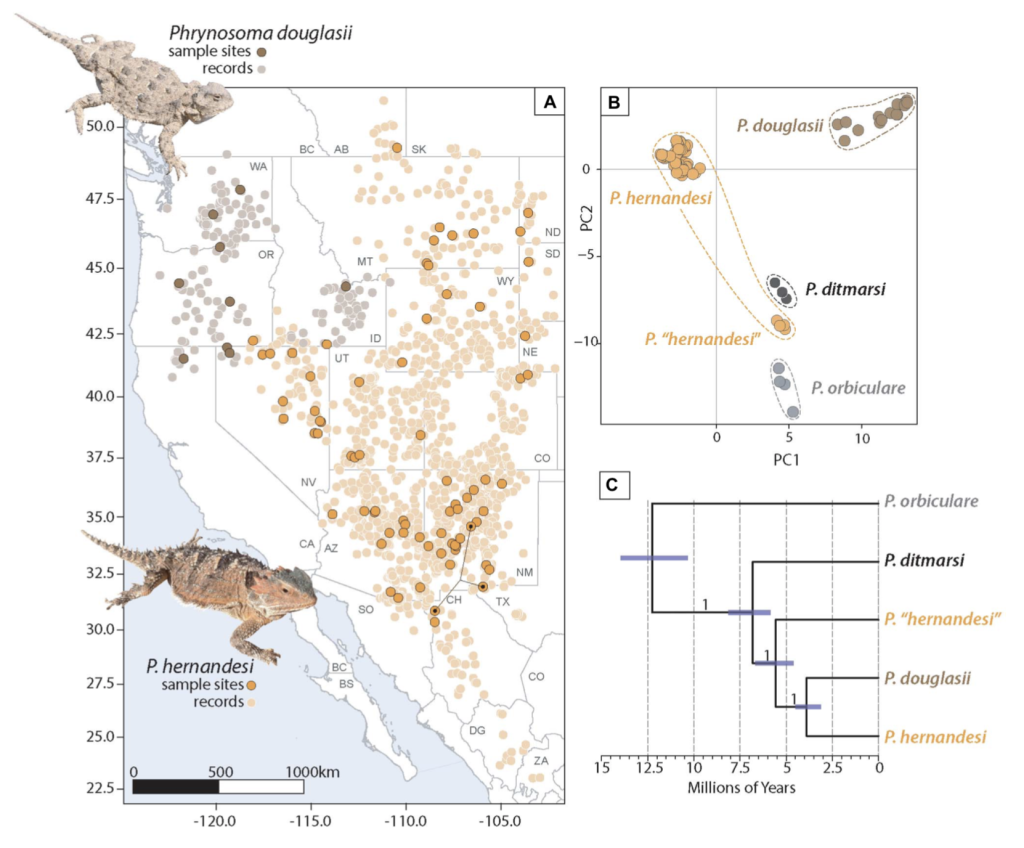
The Leaché Lab at the University of Washington in Seattle conducts research at the intersections of evolutionary biology, systematics, and biodiversity science. Based in the Department of Biology & Burke Museum, the lab focuses on the diversity, evolution, and systematics of reptiles and amphibians. We use genomic data to study natural populations, with an emphasis on applying innovative approaches to questions in phylogenetics, phylogeography, and species delimitation.