Demo
Welcome
This assumes familiarity with R, dynamical systems & malaria.
To get started, install ramp.xds from the GitHub repository using devtools::install_github():
Then load it:
A Model
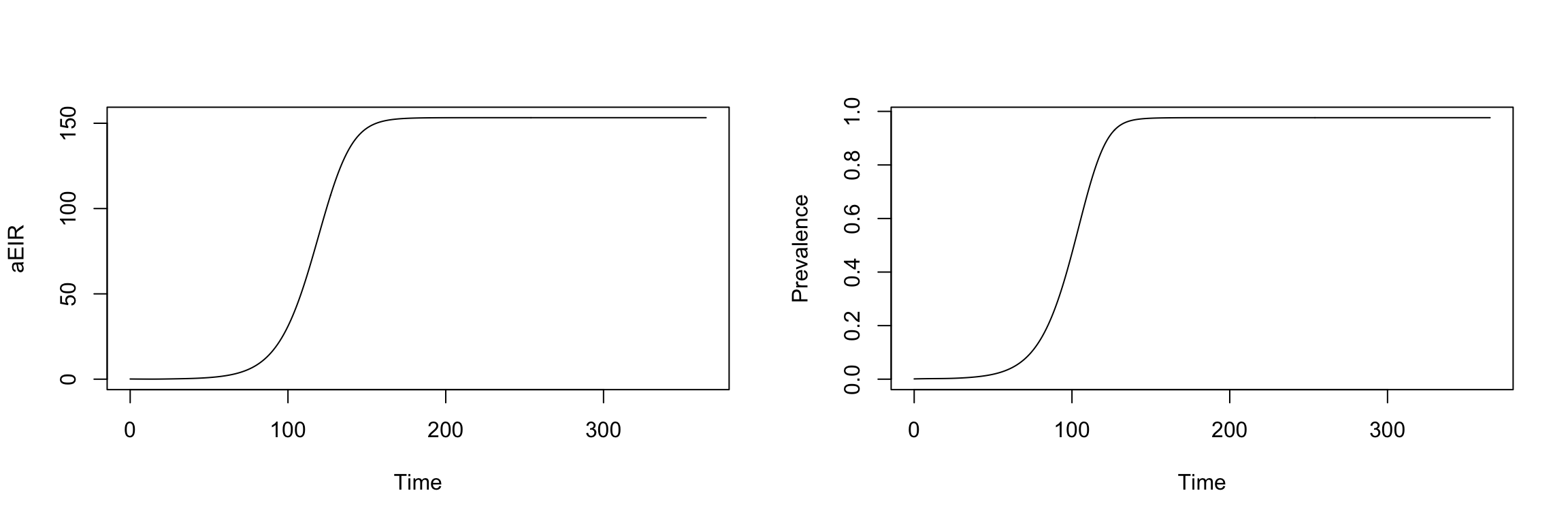
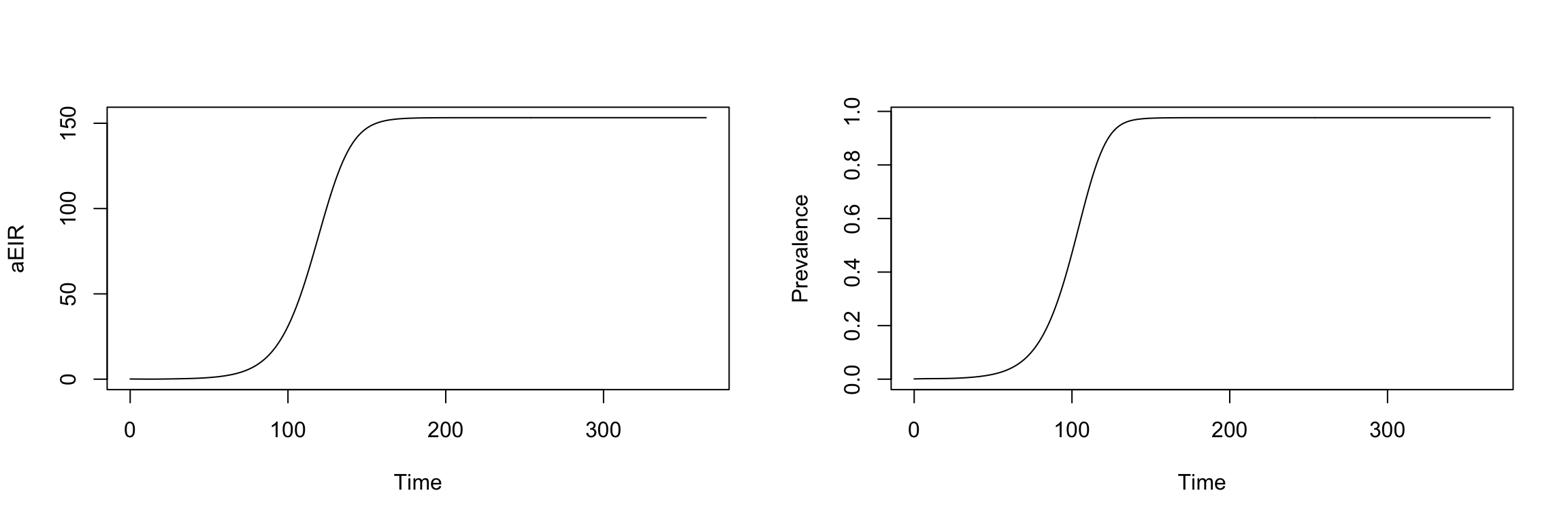
In ramp.xds, it’s easy to build, solve, and visualize malaria models, formulated as dynamical systems:
The xds Object
The object that was returned by xds_setup(), that we named demo_mod, is a compound list called an xds object. Three text strings define the class of each dynamic module:
Setup:: Defaults
xds_setup() supplies default values:
default dynamic modules (i.e.
SIS,macdonald, andtrivial) recreate a model published in 1982 [1];each dynamic module sets up its own default parameters & initial values;
all structural parameters are set to \(1\) (i.e., the number of patches, aquatic habitats, host strata, host species, & vector species)
In a few slides, we’ll describe how to change the defaults.
The xds Object : X module
The X component – a dynamical module describing infection dynamics & immunity in humans or other vertebrate hosts – is dispatched by Xpar[[i]]. Basic setup sets up only the first host species:
The xds Object : MYZ module
The MYZ component defines adult mosquito ecology and infection dynamics for each vector species, stored in MYZpar[[i]]:
The xds Object : L module
The L component defines aquatic mosquito population dynamics for each vector species, stored in Lpar[[i]]:
The trivial module is a function that emergence rate of adult mosquitoes, \(\Lambda(t).\) Here, the scaling parameter is called Lambda
’## Trivial modules & Forcing
To implement plug-and-play modularity, each component has a trivial module. All trivial modules return a function of time:
\[s \times F_S(t) \times F_T(t)\]
\(s\) is a scale parameter named in context (e.g.
Lambda)\(F_S(t)\) is the seasonal pattern, returned by
F_season\(F_T(t)\) is an inter-annual trend, returned by
F_trend
Some function families can be set up by make_function
Setup :: xds_setup
Setting up basic features is easy using xds_setup_*()
xds_setup()can be used for most situationsxds_setup_cohort()sets up a human / vertebrate host model forced by \(F_{E}(a,t),\) a function that returns the daily EIR as a function of age and time.Other
xds_setup_*()functions streamline special cases and define aframefor solving, described in the documentation
Setup :: Dynamic Modules
Dynamic modules are configured by passing:
Xname = "model_name"MYZname = "model_name"Lname = "model_name"
The values in a named list overwrite default parameters & initial values:
Xopts = list(X=1, r=1/180)
MYZopts = list(M=1, g=1/5)Lopts = list(L=1, psi = 1/5)
Setup :: Structural Elements
xds_setup() sets up a model with one host and one vector species:
nPatchesis the number of spatial units for adult mosquitoesmembershipindexes the patch membership of each habitat, passed as a vector where \(\mbox{max}(\)membership\()\leq\)nPatchesandnHabitats\(=\mbox{length}(\)membership\()\)HPopis the population size of the strata,residenceindexes the patches where the humans reside, and
nStrata\(=\mbox{length}(\)HPop\()= \mbox{length}(\)residence\().\)
Setup :: Spatial Dynamics
Solving
It’s also easy to visualize the outputs: