Lutz Lab
Department of Bioengineering, University of
Washington
Home Research Publications People News Contact
Raman nanoparticle probes for multiplexed protein detection (prior work with Intel & Beatrice Knudsen, FHCRC)
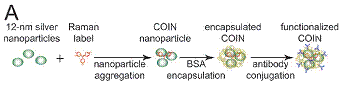 The
Intel team created composite organic-inorganic nanoparticles
(COINs) that use sliver nanoparticle clusters to
create high intensity (surface-enhanced) Raman emission of an embedded
signature molecule. COINs are similar to fluorescent reporters except that
their emission is a fingerprint of distinct spectral peaks, thus they are
well-suited for multiplexing. Many other groups have created Raman nanoparticles with similar features (often based on
adsorption of reporter molecules to gold nanoparticles).
COINs and other Raman nanoparticles can be
functionalized with binding molecules (antibodies, nucleic acid probes) for
bioassays. As a Research Scientist at Intel, I worked at the Fred Hutchinson
Cancer Research Center to apply COINs developed by the Intel team for
multiplexed detection of cancer biomarkers in tissue specimens.
The
Intel team created composite organic-inorganic nanoparticles
(COINs) that use sliver nanoparticle clusters to
create high intensity (surface-enhanced) Raman emission of an embedded
signature molecule. COINs are similar to fluorescent reporters except that
their emission is a fingerprint of distinct spectral peaks, thus they are
well-suited for multiplexing. Many other groups have created Raman nanoparticles with similar features (often based on
adsorption of reporter molecules to gold nanoparticles).
COINs and other Raman nanoparticles can be
functionalized with binding molecules (antibodies, nucleic acid probes) for
bioassays. As a Research Scientist at Intel, I worked at the Fred Hutchinson
Cancer Research Center to apply COINs developed by the Intel team for
multiplexed detection of cancer biomarkers in tissue specimens.
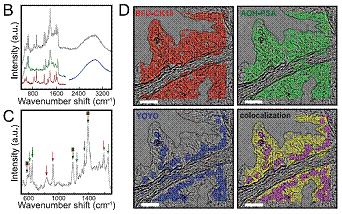 We
developed plate assays for multiplexed detection and showed that COIN signals
could be extracted in multiplexed assays on tissues. The figure at left shows
detection of three targets in a formalin-fixed paraffin-embedded prostate
tissue section. COINs targeted prostate specific antigen (PSA) and
cytokeritin-18 (CK18), and a DNA-intercalating fluorescent dye (YOYO) marked
DNA in cell nuclei. The signals from each probe were extracted from spectra
recorded at each point in a rastered image to create
false-color images of each target. In this work (ACS Nano) and other work (J. Histochem. Cytochem.), we characterized performance of labeling
and signal separation. We also showed that signals from four COINs could be
separated quantitatively within a small fraction of the visible spectrum,
suggesting that a dozen or more probes could be used for multiplexing
(potentially more than fluorophores or quantum dots).
We
developed plate assays for multiplexed detection and showed that COIN signals
could be extracted in multiplexed assays on tissues. The figure at left shows
detection of three targets in a formalin-fixed paraffin-embedded prostate
tissue section. COINs targeted prostate specific antigen (PSA) and
cytokeritin-18 (CK18), and a DNA-intercalating fluorescent dye (YOYO) marked
DNA in cell nuclei. The signals from each probe were extracted from spectra
recorded at each point in a rastered image to create
false-color images of each target. In this work (ACS Nano) and other work (J. Histochem. Cytochem.), we characterized performance of labeling
and signal separation. We also showed that signals from four COINs could be
separated quantitatively within a small fraction of the visible spectrum,
suggesting that a dozen or more probes could be used for multiplexing
(potentially more than fluorophores or quantum dots).
A generally useful outcome from this work was development of a simple spectral deconvolution algorithm that allowed separation of multiplexed Raman probe signals in the presence of unknown sample autofluorescence. Since Raman emission is characterized by sharp peaks, while autofluorescence is characterized by broad spectral background, these disparate signals can be robustly separated. We performed spectral deconvolution using reference spectra for the Raman probes, a representative autofluorescence background spectrum to account for expected background, as well as a free-fitting polynomial (or other slowly-varying function) to account for unknown background signals. All of these components were allowed to vary during the spectral fitting, and the shape of the free-fitting polynomial was also allowed to vary. This is distinctly different from baseline subtraction, which can artificially fit unknown features and corrupt the extraction of probe signals.
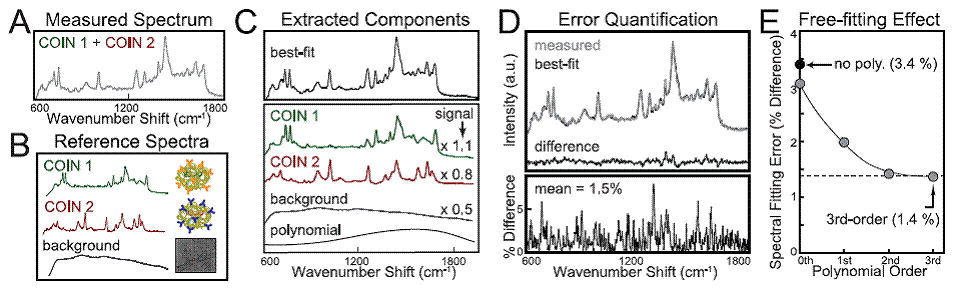
The algorithm is trivial and fast to implement, and the result is a robust separation of probe signals from background, with each component quantified. Other groups have successfully uses single peak heights to extract probe signals; I have not done a head to head comparison, but I expect that quantification would be improve by using full spectral signatures (especially for higher-order multiplexing, where single unique peaks cannot be identified, see ACS Nano paper). In addition, the spectral fitting method also provides a point-by-point estimate of fitting error; fitting errors of ~2% of signal were found across a wide range of assay formats (plate assays, tissue assays) demonstrating that the fitting is indeed robust to variations in multiplexing and unknown background. This approach takes advantage of the sharp spectral features of Raman proves (any Raman probe, not just COINs), but I expect that it could be applied to quantum dots as well. In our paper, we provided the framework of the fitting algorithm for Matlab.
References:
· Lutz, B., Dentinger, C. Sun, L., Nguyen, L., Zhang, J., Chmura, AJ, Allen, A., Chan, S., Knudsen, B. Spectral analysis of multiplex Raman probe signatures. ACS Nano, 2, 2306–2314 (2008). [pdf
]