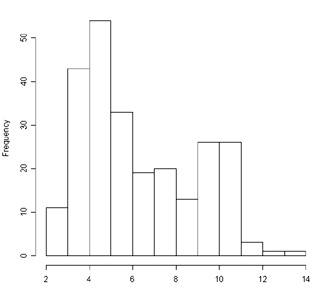
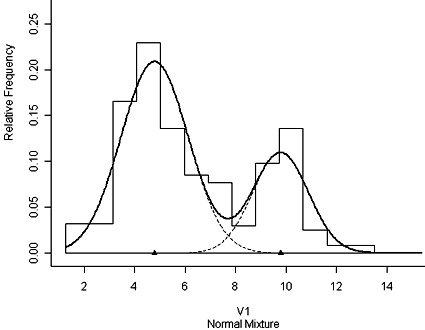
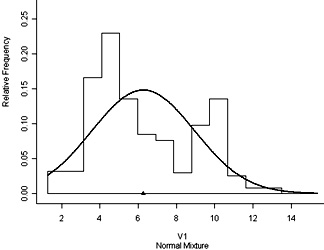
Using the R programming language you can investigate fitting a bimodal distribution to histogram data using mixdist. You must first install the package and then load it. The data used as an example is of plant height and plant weight of 250 dwarf marigold plants 42 days after sowing closely together. A histogram of the height data (shown at right) can be obtained by the R command:
> hist(Ht.data)
The scale of the x axis is centimeters and frequency is number of plants. The function hist calculates the number of bins into which values are sorted. This can be regulated using the breaks parameter of that function.