|
Baneyx BioNano Laboratories.
|
| ..Molecular Chaperones |
|
..The DnaK-DnaJ-GrpE system
|
DnaK (Hsp70) is a 69-kDa, monomeric, two-domain protein that plays a central role in de novo protein folding, host protein refolding, translocation and in the general management of the deleterious effects of stress. The DnaK N-terminus domain is responsible for its ATPase activity while the C-terminus domain is involved in substrate binding. The substrate specificity of DnaK has been inferred from the crystal structure of its C-terminal domain and by assessing the ability of the chaperone to bind to immobilized random peptide libraries. DnaK was found to exhibit a preference for heptameric stretches of amino acids composed of a 4-5 residues-long hydrophobic core enriched in leucines and flanked by basic residues. Statistical analyses indicate that these motifs are quite common, occurring every 36 residues on the average protein. It is therefore not surprising that DnaK binds to a large number of structurally and functionally unrelated proteins provided that they are in a nonnative (partially unfolded) state. However, some sites are preferred as binding constants can vary between 5 nM and 5 µM.
To properly function in vivo, DnaK requires the assistance of two additional cofactors, DnaJ (Hsp40) and GrpE. DnaJ is a 41-kDa protein that triggers ATP-hydrolysis dependent substrate association of partially folded proteins to the substrate binding cavity of DnaK, and contains a conserved J domain that is required for its association with DnaK. DnaJ can independently bind unfolded proteins with low affinity and is believed to scan partially folded substrates in order to direct DnaK to high affinity binding sites. GrpE, a homodimer of 20-kDa subunits, triggers ADP release from DnaK and subsequent substrate release as ATP rebinds DnaK. It is generally accepted that substrate proteins ejected from DnaK either fold into a proper conformation, are recaptured by DnaK-DnaJ for additional cycles of binding and release or are transferred in a partially folded form to GroEL-GroES for subsequent folding.
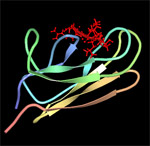
| DnaK-substrate complex. The substrate binding domain of DnaK is shown in complex with peptide NRLLLTG (red) which was selected by phage display. Structure rendered from PDB 1Q5L. |
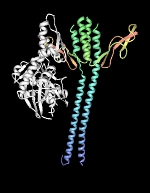
| DnaK-GrpE complex. The nucleotide exchange factor GrpE is shown in complex with the ATPase domain of DnaK (white). Structure rendered from PDB 1DKG. |
| | home | research | publications | people | covers | links | teaching | intranet |
Contact: François Baneyx, University of Washington, Department of Chemical Engineering, Box 351750, Seattle, WA Tel: 206-685-7659 Fax: 206-685-3451 E-mail: baneyx@u.washington.edu © 2008 François Baneyx - All Rights Reserved |
